4WK0
 
 | | Metal Ion and Ligand Binding of Integrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARG-GLY-ASP, ... | | Authors: | Xia, W, Springer, T.A. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Metal ion and ligand binding of integrin alpha 5 beta 1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4WJK
 
 | | Metal Ion and Ligand Binding of Integrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xia, W, Springer, T.A. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Metal ion and ligand binding of integrin alpha 5 beta 1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4WK2
 
 | | Metal Ion and Ligand Binding of Integrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xia, W, Springer, T.A. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Metal ion and ligand binding of integrin alpha 5 beta 1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4WK4
 
 | | Metal Ion and Ligand Binding of Integrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALA-CYS-ARG-GLY-ASP-GLY-TRP-CYS, ... | | Authors: | Xia, W, Springer, T.A. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Metal ion and ligand binding of integrin alpha 5 beta 1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2KDX
 
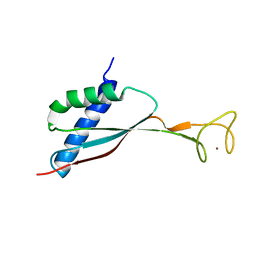 | | Solution structure of HypA protein | | Descriptor: | Hydrogenase/urease nickel incorporation protein hypA, ZINC ION | | Authors: | Xia, W, Li, H, Sze, K.-H. | | Deposit date: | 2009-01-20 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a nickel chaperone, HypA, from Helicobacter pylori reveals two distinct metal binding sites
J.Am.Chem.Soc., 131, 2009
|
|
7CJY
 
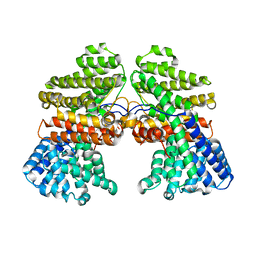 | |
6M4R
 
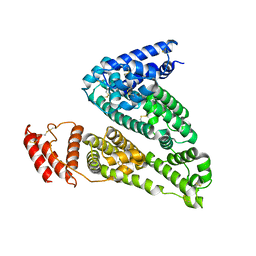 | | Structure of Human Serum Albumin | | Descriptor: | Serum albumin | | Authors: | Xiang, W, Su, J. | | Deposit date: | 2020-03-09 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure of Human Serum Albumin
To Be Published
|
|
1SOL
 
 | | A PIP2 AND F-ACTIN-BINDING SITE OF GELSOLIN, RESIDUE 150-169 (NMR, AVERAGED STRUCTURE) | | Descriptor: | GELSOLIN (150-169) | | Authors: | Xian, W, Vegners, R, Janmey, P.A, Braunlin, W.H. | | Deposit date: | 1995-09-29 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Spectroscopic studies of a phosphoinositide-binding peptide from gelsolin: behavior in solutions of mixed solvent and anionic micelles.
Biophys.J., 69, 1995
|
|
6SXX
 
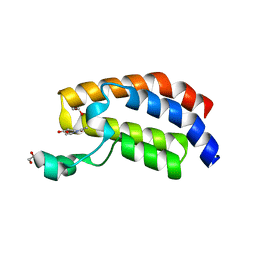 | | Crystal structure of the bromodomain of human CREBBP in complex with ACA007 | | Descriptor: | (3~{R})-1-ethanoylpyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, CREBBP | | Authors: | Xiang, W, Bedi, R.K, Sledz, P, Caflisch, A. | | Deposit date: | 2019-09-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of the bromodomain of human CREBBP in complex with ACA007
To Be Published
|
|
7VR0
 
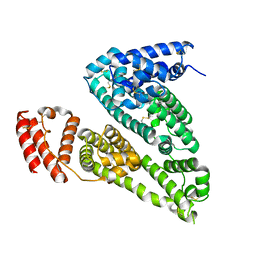 | | Human Serum Albumin | | Descriptor: | Serum albumin | | Authors: | Xiang, W, Yue, Z.L, Su, J.Y. | | Deposit date: | 2021-10-21 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Human Serum Albumin
To Be Published
|
|
7DJN
 
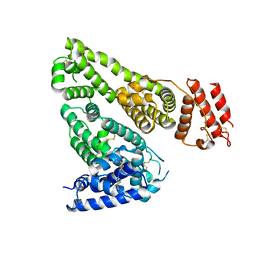 | | Human Serum Albumin | | Descriptor: | Serum albumin | | Authors: | Xiang, W, Yue, Z, Su, J. | | Deposit date: | 2020-11-20 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Human Serum Albumin
To Be Published
|
|
7FFR
 
 | |
7FFS
 
 | |
2KR7
 
 | |
6J2S
 
 | | Structure of HitA bound to gallium from Pseudomonas aeruginosa | | Descriptor: | Ferric iron-binding periplasmic protein, GALLIUM (III) ION, PHOSPHATE ION | | Authors: | Guo, Y, Li, H.Y, Sun, H.Z, Xia, W. | | Deposit date: | 2019-01-02 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.73005116 Å) | | Cite: | Identification and Characterization of a Metalloprotein Involved in Gallium Internalization in Pseudomonas aeruginosa.
Acs Infect Dis., 5, 2019
|
|
8GQD
 
 | | Complex Structure of Arginine Kinase McsB and McsA from Staphylococcus aureus | | Descriptor: | Protein-arginine kinase, Protein-arginine kinase activator protein, ZINC ION | | Authors: | Lu, K, Luo, B, Tao, X, Li, H, Xie, Y, Zhao, Z, Xia, W, Su, Z, Mao, Z. | | Deposit date: | 2022-08-30 | | Release date: | 2024-03-06 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Complex structure and activation mechanism of arginine kinase McsB by McsA.
Nat.Chem.Biol., 2024
|
|
7D64
 
 | |
5YMV
 
 | | Crystal structure of 9-mer peptide from influenza virus in complex with BF2*1201 | | Descriptor: | ALA-VAL-LYS-GLY-VAL-GLY-THR-MET-VAL, Beta-2-microglobulin, Class I histocompatibility antigen, ... | | Authors: | Xiao, J, Xiang, W, Qi, J, Chai, Y, Liu, W.J, Gao, G.F. | | Deposit date: | 2017-10-22 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | An Invariant Arginine in Common with MHC Class II Allows Extension at the C-Terminal End of Peptides Bound to Chicken MHC Class I.
J Immunol., 201, 2018
|
|
5YMW
 
 | | Crystal structure of 8-mer peptide from Rous sarcoma virus in complex with BF2*1201 | | Descriptor: | Beta-2-microglobulin, Class I histocompatibility antigen, F10 alpha chain, ... | | Authors: | Xiao, J, Xiang, W, Qi, J, Chai, Y, Liu, W.J, Gao, G.F. | | Deposit date: | 2017-10-22 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | An Invariant Arginine in Common with MHC Class II Allows Extension at the C-Terminal End of Peptides Bound to Chicken MHC Class I.
J Immunol., 201, 2018
|
|
6DKD
 
 | | Yeast Ddi2 Cyanamide Hydratase | | Descriptor: | DNA damage-inducible protein, SULFATE ION, ZINC ION | | Authors: | Moore, S.A, Xiao, W, Li, J. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Ddi2, a highly inducible detoxifying metalloenzyme fromSaccharomyces cerevisiae.
J.Biol.Chem., 294, 2019
|
|
6DKC
 
 | | Yeast Ddi2 Cyanamide Hydratase, T157V mutant, apo structure | | Descriptor: | DNA damage-inducible protein, SULFATE ION, ZINC ION | | Authors: | Moore, S.A, Xiao, W, Li, J. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of Ddi2, a highly inducible detoxifying metalloenzyme fromSaccharomyces cerevisiae.
J.Biol.Chem., 294, 2019
|
|
6DK9
 
 | | Yeast Ddi2 Cyanamide Hydratase | | Descriptor: | DNA damage-inducible protein, SULFATE ION, ZINC ION | | Authors: | Moore, S.A, Xiao, W, Li, J. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Ddi2, a highly inducible detoxifying metalloenzyme fromSaccharomyces cerevisiae.
J.Biol.Chem., 294, 2019
|
|
6DKA
 
 | | Yeast Ddi2 Cyanamide Hydratase | | Descriptor: | CYANAMIDE, DNA damage-inducible protein, SULFATE ION, ... | | Authors: | Moore, S.A, Xiao, W, Li, J. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structure of Ddi2, a highly inducible detoxifying metalloenzyme fromSaccharomyces cerevisiae.
J.Biol.Chem., 294, 2019
|
|
1J74
 
 | | Crystal Structure of Mms2 | | Descriptor: | MMS2 | | Authors: | Moraes, T.F, Edwards, R.A, McKenna, S, Pastushok, L, Xiao, W, Glover, J.N.M, Ellison, M.J. | | Deposit date: | 2001-05-15 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the human ubiquitin conjugating enzyme complex, hMms2-hUbc13.
Nat.Struct.Biol., 8, 2001
|
|
1J7D
 
 | | Crystal Structure of hMms2-hUbc13 | | Descriptor: | MMS2, UBIQUITIN-CONJUGATING ENZYME E2-17 KDA | | Authors: | Moraes, T.F, Edwards, R.A, McKenna, S, Pashushok, L, Xiao, W, Glover, J.N.M, Ellison, M.J. | | Deposit date: | 2001-05-16 | | Release date: | 2001-08-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the human ubiquitin conjugating enzyme complex, hMms2-hUbc13.
Nat.Struct.Biol., 8, 2001
|
|
