3TOS
 
 | | Crystal Structure of CalS11, Calicheamicin Methyltransferase | | Descriptor: | 1,2-ETHANEDIOL, CalS11, GLUTAMIC ACID, ... | | Authors: | Chang, A, Aceti, D.J, Beebe, E.T, Makino, S.-I, Wrobel, R.L, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2011-09-06 | | Release date: | 2011-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of CalS11, Calicheamicin methyltransferase
To be Published
|
|
4PED
 
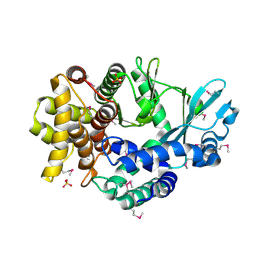 | | Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthes | | Descriptor: | Chaperone activity of bc1 complex-like, mitochondrial, SULFATE ION | | Authors: | Bingman, C.A, Smith, R, Joshi, S, Stefely, J.A, Reidenbach, A.G, Ulbrich, A, Oruganty, O, Floyd, B.J, Jochem, A, Saunders, J.M, Johnson, I.E, Wrobel, R.L, Barber, G.E, Lee, D, Li, S, Kannan, N, Coon, J.J, Pagliarini, D.J, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mitochondrial ADCK3 Employs an Atypical Protein Kinase-like Fold to Enable Coenzyme Q Biosynthesis.
Mol.Cell, 57, 2015
|
|
4RHP
 
 | | Crystal structure of human COQ9 in complex with a phospholipid, Northeast Structural Genomics Consortium Target HR5043 | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Ubiquinone biosynthesis protein COQ9, mitochondrial | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Wang, H, Lee, D, Kogan, S, Maglaqui, M, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2014-10-02 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2Q3O
 
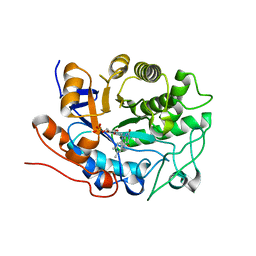 | | Ensemble refinement of the protein crystal structure of 12-oxo-phytodienoate reductase isoform 3 | | Descriptor: | 12-oxophytodienoate reductase 3, FLAVIN MONONUCLEOTIDE | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2Q3M
 
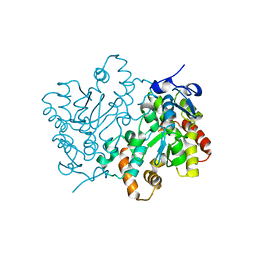 | | Ensemble refinement of the protein crystal structure of an Arabidopsis thaliana putative steroid sulphotransferase | | Descriptor: | Flavonol sulfotransferase-like, MALONIC ACID | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
1XOY
 
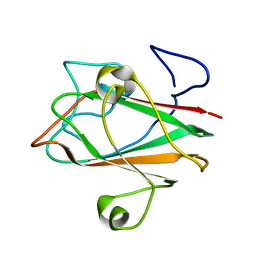 | | Solution structure of At3g04780.1, an Arabidopsis ortholog of the C-terminal domain of human thioredoxin-like protein | | Descriptor: | hypothetical protein At3g04780.1 | | Authors: | Song, J, Robert, C.T, Lee, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-07 | | Release date: | 2004-10-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of At3g04780.1-des15, an Arabidopsis thaliana ortholog of the C-terminal domain of human thioredoxin-like protein.
Protein Sci., 14, 2005
|
|
1VJI
 
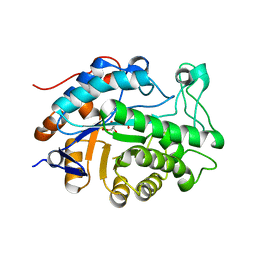 | | Gene Product of At1g76680 from Arabidopsis thaliana | | Descriptor: | 12-oxophytodienoate reductase (OPR1), FLAVIN MONONUCLEOTIDE | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | X-ray structure of Arabidopsis At1g77680, 12-oxophytodienoate reductase isoform 1.
Proteins, 61, 2005
|
|
1XRI
 
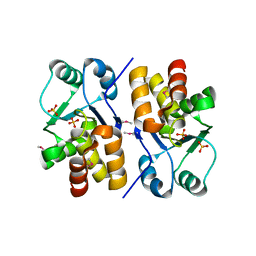 | | X-ray structure of a putative phosphoprotein phosphatase from Arabidopsis thaliana gene AT1G05000 | | Descriptor: | At1g05000, SULFATE ION | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-14 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and functional characterization of a novel phosphatase from the Arabidopsis thaliana gene locus At1g05000.
Proteins, 73, 2008
|
|
1Y0Z
 
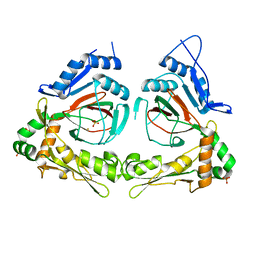 | | X-ray structure of gene product from Arabidopsis thaliana At3g21360 | | Descriptor: | FE (II) ION, SULFATE ION, protein product of AT3G21360 | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-11-16 | | Release date: | 2004-11-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure at 2.4 A resolution of the protein from gene locus At3g21360, a putative Fe(II)/2-oxoglutarate-dependent enzyme from Arabidopsis thaliana.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
2LUZ
 
 | | Solution NMR Structure of CalU16 from Micromonospora echinospora, Northeast Structural Genomics Consortium (NESG) Target MiR12 | | Descriptor: | CalU16 | | Authors: | Ramelot, T.A, Yang, Y, Lee, H, Pederson, K, Lee, D, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Wrobel, R.L, Bingman, C.A, Singh, S, Thorson, J.S, Prestegard, J.H, Montelione, G.T, Phillips Jr, G.N, Kennedy, M.A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-22 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure-Guided Functional Characterization of Enediyne Self-Sacrifice Resistance Proteins, CalU16 and CalU19.
Acs Chem.Biol., 9, 2014
|
|
2M2B
 
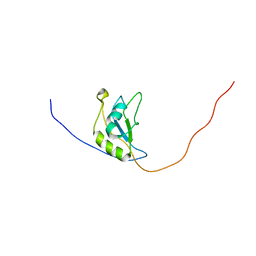 | | NMR structure of the RRM2 domain of the protein RBM10 from Homo sapiens | | Descriptor: | RNA-binding protein 10 | | Authors: | Serrano, P, Geralt, M, Dutta, S.K, Wuthrich, K, Wrobel, R.L, Makino, S, Misenhiemer, T.M, Markley, J.L, Fox, B.G, Joint Center for Structural Genomics (JCSG), Partnership for T-Cell Biology (TCELL), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2012-12-17 | | Release date: | 2013-01-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the RRM2 domain of the protein RBM10 from Homo sapiens
To be Published
|
|
2Q4A
 
 | | Ensemble refinement of the protein crystal structure of gene product from Arabidopsis thaliana At3g21360 | | Descriptor: | Clavaminate synthase-like protein At3g21360, FE (III) ION, SULFATE ION | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
1Q44
 
 | | Crystal Structure of an Arabidopsis Thaliana Putative Steroid Sulfotransferase | | Descriptor: | MALONIC ACID, Steroid Sulfotransferase | | Authors: | Phillips Jr, G.N, Smith, D.W, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-08-01 | | Release date: | 2003-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of At2g03760, a putative steroid sulfotransferase from Arabidopsis thaliana
Proteins, 57, 2004
|
|
1Q45
 
 | | 12-0xo-phytodienoate reductase isoform 3 | | Descriptor: | 12-oxophytodienoate-10,11-reductase, FLAVIN MONONUCLEOTIDE | | Authors: | Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Smith, D.W, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-08-01 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of Arabidopsis At2g06050, 12-oxophytodienoate reductase isoform 3
Proteins, 58, 2005
|
|
