8ODW
 
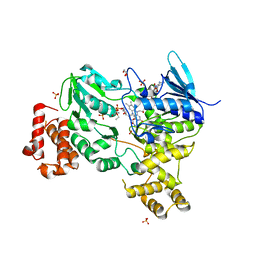 | | Crystal structure of LbmA Ox-ACP didomain in complex with NADP and ethyl glycinate from the lobatamide PKS (Gynuella sunshinyii) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Francois, R.M.M, Fraley, A.E, Piel, J, Weissman, K.J, Gruez, A. | | Deposit date: | 2023-03-09 | | Release date: | 2023-05-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Modular Oxime Formation by a trans-AT Polyketide Synthase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
4CA3
 
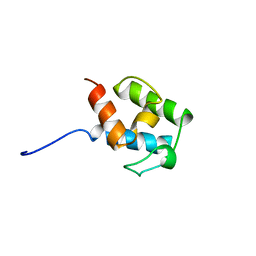 | | SOLUTION STRUCTURE OF STREPTOMYCES VIRGINIAE VIRA ACP5B | | Descriptor: | HYBRID POLYKETIDE SYNTHASE-NON RIBOSOMAL PEPTIDE SYNTHETASE | | Authors: | Davison, J, Dorival, J, Rabeharindranto, M.H, Chagot, B, Gruez, A, Weissman, K.J. | | Deposit date: | 2013-10-05 | | Release date: | 2014-06-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Insights Into the Function of Trans-Acyl Transferase Polyketide Synthases from the Saxs Structure of a Complete Module.
Chem.Sci., 2014
|
|
2JUG
 
 | |
8A7Z
 
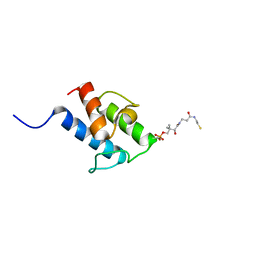 | | NMR structure of holo-acp | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Hybrid polyketide synthase-non ribosomal peptide synthetase | | Authors: | Collin, S, Weissman, K.J, Chagot, B, Gruez, A. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
8AIG
 
 | | NMR structure of holo-acp | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Hybrid non ribosomal peptide synthetase-polyketide synthase | | Authors: | Collin, S, Weissman, K.J, Chagot, B, Gruez, A. | | Deposit date: | 2022-07-26 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
8ALL
 
 | | NMR structure of holo-acp | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Hybrid non ribosomal peptide synthetase-polyketide synthase | | Authors: | Collin, S, Weissman, K.J, Chagot, B, Gruez, A. | | Deposit date: | 2022-08-01 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
6F7L
 
 | | Crystal structure of LkcE R326Q mutant in complex with its substrate | | Descriptor: | ACETATE ION, Amine oxidase LkcE, CALCIUM ION, ... | | Authors: | Dorival, J, Risser, F, Jacob, C, Collin, S, Drager, G, Kirschning, A, Paris, C, Chagot, B, Gruez, A, Weissman, K.J. | | Deposit date: | 2017-12-11 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into a dual function amide oxidase/macrocyclase from lankacidin biosynthesis.
Nat Commun, 9, 2018
|
|
6FJH
 
 | | Crystal structure of the seleniated LkcE from Streptomyces rochei | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LkcE, OXYGEN MOLECULE, ... | | Authors: | Dorival, J, Risser, F, Jacob, C, Collin, S, Drager, G, Kirschning, A, Paris, C, Chagot, B, Gruez, A, Weissman, K.J. | | Deposit date: | 2018-01-22 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Insights into a dual function amide oxidase/macrocyclase from lankacidin biosynthesis.
Nat Commun, 9, 2018
|
|
6F32
 
 | | Crystal structure of a dual function amine oxidase/cyclase in complex with substrate analogues | | Descriptor: | (2~{R},3~{R})-2,3-bis(oxidanyl)-~{N},~{N}'-dipropyl-butanediamide, ACETATE ION, Amine oxidase LkcE, ... | | Authors: | Dorival, J, Risser, F, Jacob, C, Collin, S, Drager, G, Kirschning, A, Paris, C, Chagot, B, Gruez, A, Weissman, K.J. | | Deposit date: | 2017-11-27 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into a dual function amide oxidase/macrocyclase from lankacidin biosynthesis.
Nat Commun, 9, 2018
|
|
6F7V
 
 | | Crystal structure of LkcE E64Q mutant in complex with LC-KA05 | | Descriptor: | CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Dorival, J, Risser, F, Jacob, C, Collin, S, Drager, G, Kirschning, A, Paris, C, Chagot, B, Gruez, A, Weissman, K.J. | | Deposit date: | 2017-12-12 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Insights into a dual function amide oxidase/macrocyclase from lankacidin biosynthesis.
Nat Commun, 9, 2018
|
|
1PZQ
 
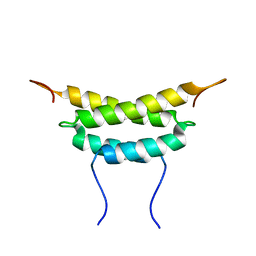 | | Structure of fused docking domains from the erythromycin polyketide synthase (DEBS), a model for the interaction between DEBS 2 and DEBS 3: The A domain | | Descriptor: | Erythronolide synthase | | Authors: | Broadhurst, R.W, Nietlispach, D, Wheatcroft, M.P, Leadlay, P.F, Weissman, K.J. | | Deposit date: | 2003-07-14 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of docking domains in modular polyketide synthases.
Chem.Biol., 10, 2003
|
|
1PZR
 
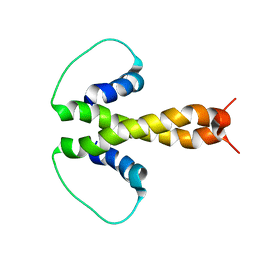 | | Structure of fused docking domains from the erythromycin polyketide synthase (DEBS), a model for the interaction between DEBS2 and DEBS3: the B domain | | Descriptor: | Erythronolide synthase | | Authors: | Broadhurst, R.W, Nietlispach, D, Wheatcroft, M.P, Leadlay, P.F, Weissman, K.J. | | Deposit date: | 2003-07-14 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of docking domains in modular polyketide synthases.
Chem.Biol., 10, 2003
|
|
8RAJ
 
 | | NMR structure of PKS docking domains | | Descriptor: | Beta-ketoacyl synthase, Trimethylamine monooxygenase | | Authors: | Scat, S, Weissman, K.J, Chagot, B. | | Deposit date: | 2023-12-01 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-17 | | Method: | SOLUTION NMR | | Cite: | Insights into docking in megasynthases from the investigation of the toblerol trans -AT polyketide synthase: many alpha-helical means to an end.
Rsc Chem Biol, 5, 2024
|
|
2MF4
 
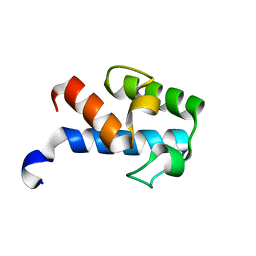 | | 1H, 13C, 15N chemical shift assignments of Streptomyces virginiae VirA acp5a | | Descriptor: | Hybrid polyketide synthase-non ribosomal peptide synthetase | | Authors: | Davison, J, Dorival, J, Rabeharindranto, M.H, Mazon, H, Chagot, B, Gruez, A, Weissman, K.J. | | Deposit date: | 2013-10-05 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR assignements of ACP5a
To be Published
|
|
2N5D
 
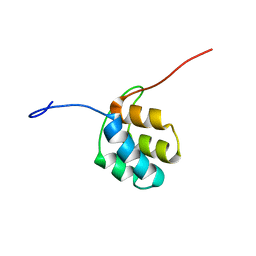 | | NMR structure of PKS domains | | Descriptor: | fusion protein of two PKS domains | | Authors: | Dorival, J, Annaval, T, Risser, F, Collin, S, Roblin, P, Jacob, C, Gruez, A, Chagot, B, Weissman, K.J. | | Deposit date: | 2015-07-14 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of Intersubunit Communication in the Virginiamycin trans-Acyl Transferase Polyketide Synthase.
J.Am.Chem.Soc., 138, 2016
|
|
8AHQ
 
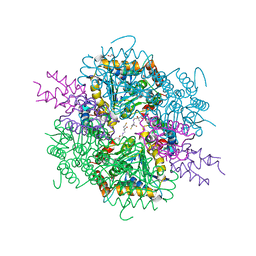 | | VirD/holo-ACP5b of Streptomyces virginiae complex | | Descriptor: | 1,2-ETHANEDIOL, 4'-PHOSPHOPANTETHEINE, CHLORIDE ION, ... | | Authors: | Collin, S, Gruez, A. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
8AHZ
 
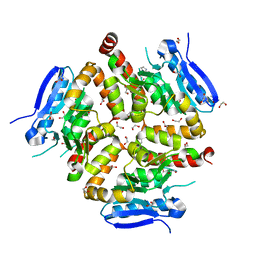 | | Native VirD of Streptomyces virginiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Enoyl-CoA hydratase, ... | | Authors: | Collin, S, Gruez, A. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
6TDM
 
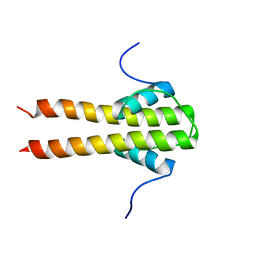 | | Bam_5920cDD 5919nDD docking domains | | Descriptor: | Beta-ketoacyl synthase,Beta-ketoacyl synthase | | Authors: | Risser, F, Chagot, B. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: Docking in the enacyloxin IIa polyketide synthase.
J.Struct.Biol., 212, 2020
|
|
6TDD
 
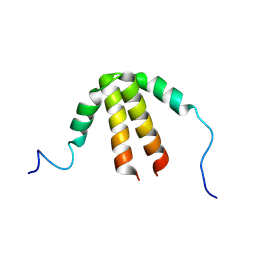 | | Bam_5924 docking domain | | Descriptor: | Beta-ketoacyl synthase | | Authors: | Risser, F, Chagot, B. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: Docking in the enacyloxin IIa polyketide synthase.
J.Struct.Biol., 212, 2020
|
|
6TDN
 
 | | Bam_5925cDD 5924nDD docking domains | | Descriptor: | Beta-ketoacyl synthase,Beta-ketoacyl synthase | | Authors: | Risser, F, Chagot, B. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: Docking in the enacyloxin IIa polyketide synthase.
J.Struct.Biol., 212, 2020
|
|
