1QTX
 
 | |
1QS7
 
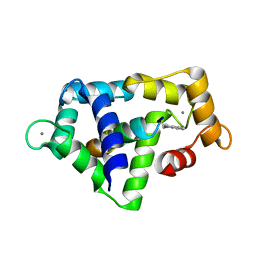 | |
1VRK
 
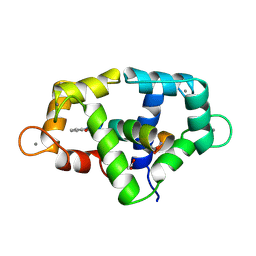 | |
3S1A
 
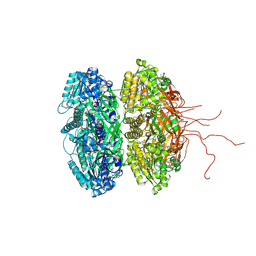 | | Crystal structure of the phosphorylation-site double mutant S431E/T432E of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Williams, D.W, Rossi, G, Weigand, S, Mori, T, Johnson, C.H, Stewart, P.L, Egli, M. | | Deposit date: | 2011-05-14 | | Release date: | 2011-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Combined SAXS/EM Based Models of the S. elongatus Post-Translational Circadian Oscillator and its Interactions with the Output His-Kinase SasA.
Plos One, 6, 2011
|
|
1D8F
 
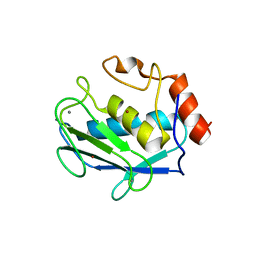 | | CRYSTAL STRUCTURE OF MMP3 COMPLEXED WITH A PIPERAZINE BASED INHIBITOR. | | Descriptor: | CALCIUM ION, N-HYDROXY-1-(4-METHOXYPHENYL)SULFONYL-4-BENZYLOXYCARBONYL-PIPERAZINE-2-CARBOXAMIDE, STROMELYSIN-1 PRECURSOR, ... | | Authors: | Cheng, M.Y, De, B, Pikul, S, Almstead, N.G, Natchus, M.G, Anastasio, M.V, McPhail, S.J, Snider, C.E, Taiwo, Y.O, Chen, L.Y. | | Deposit date: | 1999-10-22 | | Release date: | 2000-10-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of piperazine-based matrix metalloproteinase inhibitors.
J.Med.Chem., 43, 2000
|
|
4R9M
 
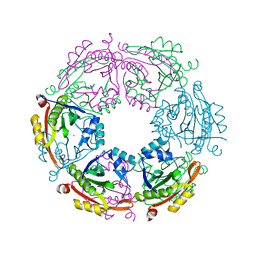 | | Crystal structure of spermidine N-acetyltransferase from Escherichia coli | | Descriptor: | MAGNESIUM ION, Spermidine N(1)-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Grimshaw, S, Wolfe, A.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-05 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Analysis of crystalline and solution states of ligand-free spermidine N-acetyltransferase (SpeG) from Escherichia coli.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5CNP
 
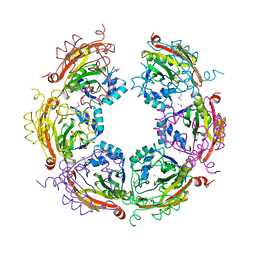 | | X-ray crystal structure of Spermidine n1-acetyltransferase from Vibrio cholerae. | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, Spermidine N(1)-acetyltransferase | | Authors: | Osipiuk, J, VOLKART, L, MOY, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-07-17 | | Release date: | 2015-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Substrate-Induced Allosteric Change in the Quaternary Structure of the Spermidine N-Acetyltransferase SpeG.
J.Mol.Biol., 427, 2015
|
|
4JLY
 
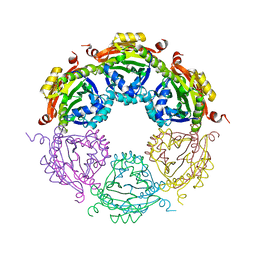 | | Dodecameric structure of spermidine N-acetyltransferase from Vibrio cholerae | | Descriptor: | SULFATE ION, Spermidine n1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-13 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.882 Å) | | Cite: | Substrate-Induced Allosteric Change in the Quaternary Structure of the Spermidine N-Acetyltransferase SpeG.
J.Mol.Biol., 427, 2015
|
|
6ELL
 
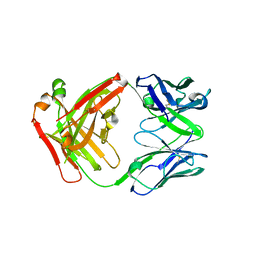 | | FAB Fragment. AbVance: Increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery | | Descriptor: | fAB heavy chain, fAB light chain | | Authors: | Benz, J, Weigand, S, Dengl, S, Schlothauer, T, Auer, J, Ehler, A, Kettenberger, H, Lorenz, S, Hirschheydt, T, Georges, G. | | Deposit date: | 2017-09-29 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AbVance: increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery
To Be Published
|
|
6EMJ
 
 | | FAB Fragment. AbVance: Increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery | | Descriptor: | SODIUM ION, fAB heavy chain, fAb light chain | | Authors: | Benz, J, Weigand, S, Dengl, S, Schlothauer, T, Auer, J, Ehler, A, Kettenberger, H, Lorenz, S, Hirschheydt, T, Georges, G. | | Deposit date: | 2017-10-02 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AbVance: increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery
To Be Published
|
|
6CY6
 
 | | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Escherichia coli in complex with tris(hydroxymethyl)aminomethane. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-04 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Analysis of crystalline and solution states of ligand-free spermidine N-acetyltransferase (SpeG) from Escherichia coli.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4YGO
 
 | | Dodecameric structure of spermidine N-acetyltransferase from Vibrio cholerae in intermediate state | | Descriptor: | CALCIUM ION, METHANOL, Spermidine n1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-02-26 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate-Induced Allosteric Change in the Quaternary Structure of the Spermidine N-Acetyltransferase SpeG.
J.Mol.Biol., 427, 2015
|
|
5LVX
 
 | | Crystal structure of glucocerebrosidase with an inhibitory quinazoline modulator | | Descriptor: | 11-[(2~{R})-2-[(2-pyridin-3-ylquinazolin-4-yl)amino]-2,3-dihydro-1~{H}-inden-5-yl]undec-10-ynoic acid, 11-[(2~{S})-2-[(2-pyridin-3-ylquinazolin-4-yl)amino]-2,3-dihydro-1~{H}-inden-5-yl]undec-10-ynoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zheng, J, Chen, L, Skinner, O.S, Lansbury, P, Skerlj, R, Mrosek, M, Heunisch, U, Krapp, S, Weigand, S, Charrow, J, Schwake, M, Kelleher, N.L, Silverman, R.B, Krainc, D. | | Deposit date: | 2016-09-14 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | beta-Glucocerebrosidase Modulators Promote Dimerization of beta-Glucocerebrosidase and Reveal an Allosteric Binding Site.
J. Am. Chem. Soc., 140, 2018
|
|
6ELJ
 
 | | FAB Fragment. AbVance: Increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery | | Descriptor: | fAB heavy chain, fAB light chain | | Authors: | Benz, J, Weigand, S, Dengl, S, Schlothauer, T, Auer, J, Ehler, A, Kettenberger, H, Lorenz, S, Hirschheydt, T, Georges, G. | | Deposit date: | 2017-09-29 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AbVance: increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery
To Be Published
|
|
6ELE
 
 | | FAB Fragment. AbVance: Increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, fAB heavy chain, ... | | Authors: | Benz, J, Weigand, S, Dengl, S, Schlothauer, T, Auer, J, Ehler, A, Kettenberger, H, Lorenz, S, Hirschheydt, T, Georges, G. | | Deposit date: | 2017-09-28 | | Release date: | 2017-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | AbVance: increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery
To Be Published
|
|
