5NPA
 
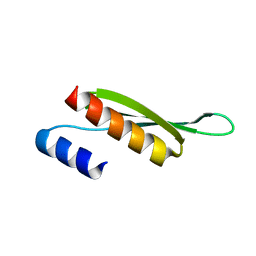 | | Solution structure of Drosophila melanogaster Loquacious dsRBD2 | | Descriptor: | Loquacious | | Authors: | Tants, J.-N, Fesser, S, Kern, T, Stehle, R, Geerlof, A, Wunderlich, C, Boettcher, R, Kunzelmann, S, Lange, O, Kreutz, C, Foerstemann, K, Sattler, M. | | Deposit date: | 2017-04-16 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for asymmetry sensing of siRNAs by the Drosophila Loqs-PD/Dcr-2 complex in RNA interference.
Nucleic Acids Res., 45, 2017
|
|
5NPG
 
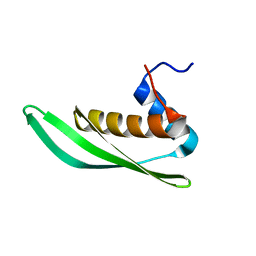 | | Solution structure of Drosophila melanogaster Loquacious dsRBD1 | | Descriptor: | Loquacious, isoform F | | Authors: | Tants, J.-N, Fesser, S, Kern, T, Stehle, R, Geerlof, A, Wunderlich, C, Hartlmuller, C, Boettcher, R, Kunzelmann, S, Lange, O, Kreutz, C, Foerstemann, K, Sattler, M. | | Deposit date: | 2017-04-16 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for asymmetry sensing of siRNAs by the Drosophila Loqs-PD/Dcr-2 complex in RNA interference.
Nucleic Acids Res., 45, 2017
|
|
5JS7
 
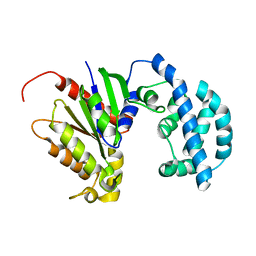 | | Structural model of a apo G-protein alpha subunit determined with NMR residual dipolar couplings and SAXS | | Descriptor: | Guanine nucleotide-binding protein G(i) subunit alpha-1 | | Authors: | Goricanec, D, Stehle, R, Grigoriu, S, Wagner, G, Hagn, F. | | Deposit date: | 2016-05-07 | | Release date: | 2016-06-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational dynamics of a G-protein alpha subunit is tightly regulated by nucleotide binding.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JS8
 
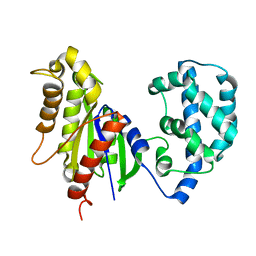 | | Structural Model of a Protein alpha subunit in complex with GDP obtained with SAXS and NMR residual couplings | | Descriptor: | Guanine nucleotide-binding protein G(i) subunit alpha-1 | | Authors: | Goricanec, D, Stehle, R, Grigoriu, S, Wagner, G, Hagn, F. | | Deposit date: | 2016-05-07 | | Release date: | 2016-06-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational dynamics of a G-protein alpha subunit is tightly regulated by nucleotide binding.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6TR0
 
 | | Solution structure of U2AF2 RRM1,2 | | Descriptor: | Splicing factor U2AF 65 kDa subunit | | Authors: | Kang, H.-S, Sattler, M. | | Deposit date: | 2019-12-17 | | Release date: | 2020-05-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | An autoinhibitory intramolecular interaction proof-reads RNA recognition by the essential splicing factor U2AF2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5JU7
 
 | | DNA BINDING DOMAIN OF E.COLI CADC | | Descriptor: | Transcriptional activator CadC, ZINC ION | | Authors: | Janowski, R, Schlundt, A, Sattler, M, Niessing, D. | | Deposit date: | 2016-05-10 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-function analysis of the DNA-binding domain of a transmembrane transcriptional activator.
Sci Rep, 7, 2017
|
|
8B8S
 
 | | Solution structure of tandem RRM1 and RRM2 domains of yeast NPL3 | | Descriptor: | Serine/arginine (SR)-type shuttling mRNA binding protein NPL3 | | Authors: | Kachariya, N, Sattler, M, Keil, P, Strasser, K. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Npl3 functions in mRNP assembly by recruitment of mRNP components to the transcription site and their transfer onto the mRNA.
Nucleic Acids Res., 51, 2023
|
|
5M0H
 
 | |
5M0J
 
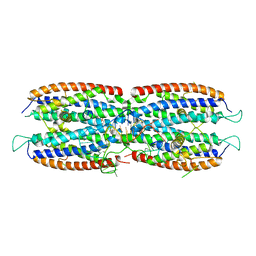 | | Crystal structure of the cytoplasmic complex with She2p, She3p, and the ASH1 mRNA E3-localization element | | Descriptor: | ASH1 E3 (28 nt-loop), MAGNESIUM ION, SWI5-dependent HO expression protein 2,SWI5-dependent HO expression protein 3 | | Authors: | Edelmann, F.T, Janowski, R, Niessing, D. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular architecture and dynamics of ASH1 mRNA recognition by its mRNA-transport complex.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5M0I
 
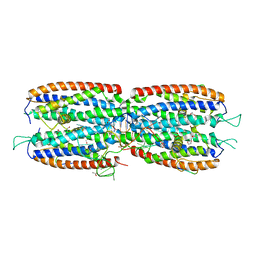 | | Crystal structure of the nuclear complex with She2p and the ASH1 mRNA E3-localization element | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ASH1-E3 element, ... | | Authors: | Edelmann, F.T, Janowski, R, Niessing, D. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Molecular architecture and dynamics of ASH1 mRNA recognition by its mRNA-transport complex.
Nat. Struct. Mol. Biol., 24, 2017
|
|
4QI0
 
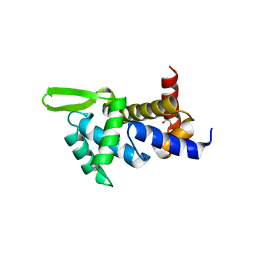 | | X-ray structure of the ROQ domain from murine Roquin-1 | | Descriptor: | 1,2-ETHANEDIOL, Roquin-1 | | Authors: | Janowski, R, Schlundt, A, Sattler, M, Niessing, D. | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis for RNA recognition in roquin-mediated post-transcriptional gene regulation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
5O3J
 
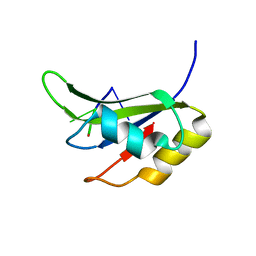 | | Crystal structure of TIA-1 RRM2 in complex with RNA | | Descriptor: | Nucleolysin TIA-1 isoform p40, RNA (5'-R(P*UP*UP*C)-3') | | Authors: | Sonntag, M, Jagtap, P.K.A, Hennig, J, Sattler, M. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Segmental, Domain-Selective Perdeuteration and Small-Angle Neutron Scattering for Structural Analysis of Multi-Domain Proteins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5O2V
 
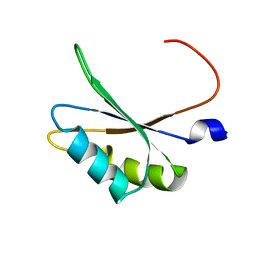 | | NMR structure of TIA-1 RRM1 domain | | Descriptor: | Nucleolysin TIA-1 isoform p40 | | Authors: | Jagtap, P.K.A. | | Deposit date: | 2017-05-22 | | Release date: | 2017-06-28 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Segmental, Domain-Selective Perdeuteration and Small-Angle Neutron Scattering for Structural Analysis of Multi-Domain Proteins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
4UJ5
 
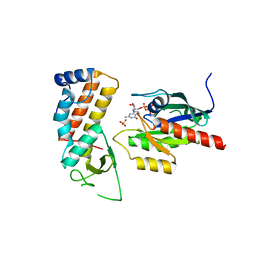 | | Crystal structure of human Rab11-Rabin8-FIP3 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, RAB-3A-INTERACTING PROTEIN, ... | | Authors: | Vetter, M, Lorentzen, E. | | Deposit date: | 2015-04-08 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structure of Rab11-Fip3-Rabin8 Reveals Simultaneous Binding of Fip3 and Rabin8 Effectors to Rab11.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UJ4
 
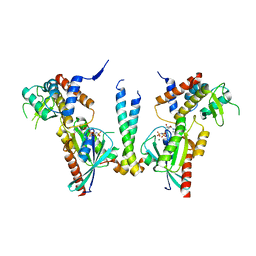 | | Crystal structure of human Rab11-Rabin8-FIP3 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Rab-3A-interacting protein, ... | | Authors: | Vetter, M, Lorentzen, E. | | Deposit date: | 2015-04-08 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structure of Rab11-FIP3-Rabin8 reveals simultaneous binding of FIP3 and Rabin8 effectors to Rab11.
Nat. Struct. Mol. Biol., 22, 2015
|
|
4UJ3
 
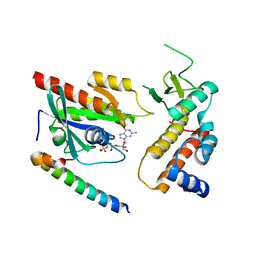 | | Crystal structure of human Rab11-Rabin8-FIP3 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, RAB-3A-INTERACTING PROTEIN, ... | | Authors: | Vetter, M, Lorentzen, E. | | Deposit date: | 2015-04-08 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Rab11-Fip3-Rabin8 Reveals Simultaneous Binding of Fip3 and Rabin8 Effectors to Rab11.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4QI2
 
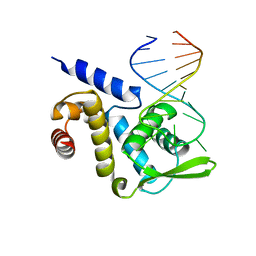 | | X-ray structure of the ROQ domain from murine Roquin-1 in complex with a 23-mer Tnf-CDE RNA | | Descriptor: | RNA (5'-R(*AP*CP*AP*UP*GP*UP*UP*UP*UP*CP*UP*GP*UP*GP*AP*AP*AP*AP*CP*GP*GP*AP*G)-3'), Roquin-1 | | Authors: | Janowski, R, Schlundt, A, Sattler, M, Niessing, D. | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for RNA recognition in roquin-mediated post-transcriptional gene regulation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
7PDV
 
 | |
7PCV
 
 | |
6GD2
 
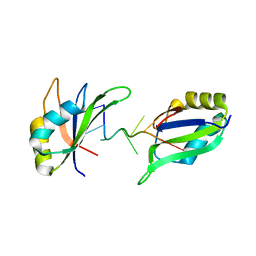 | | Structure of HuR RRM3 in complex with RNA | | Descriptor: | ELAV-like protein 1, RNA (5'-R(P*UP*UP*UP*AP*UP*UP*U)-3') | | Authors: | Pabis, M, Sattler, M. | | Deposit date: | 2018-04-21 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs.
Nucleic Acids Res., 47, 2019
|
|
6GD3
 
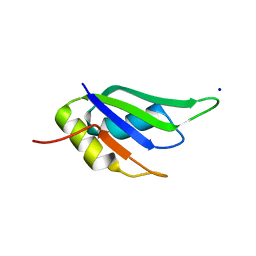 | | Structure of HuR RRM3 in complex with RNA (UAUUUA) | | Descriptor: | ELAV-like protein 1, RNA (5'-R(P*UP*AP*UP*UP*UP*A)-3'), SODIUM ION | | Authors: | Pabis, M, Sattler, M. | | Deposit date: | 2018-04-21 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs.
Nucleic Acids Res., 47, 2019
|
|
6G2K
 
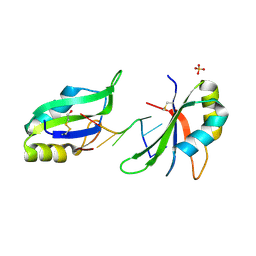 | | Structure of HuR RRM3 in complex with RNA (UUUUUU) | | Descriptor: | ELAV-like protein 1, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3'), SULFATE ION | | Authors: | Pabis, M, Sattler, M. | | Deposit date: | 2018-03-23 | | Release date: | 2018-10-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs.
Nucleic Acids Res., 47, 2019
|
|
6GD1
 
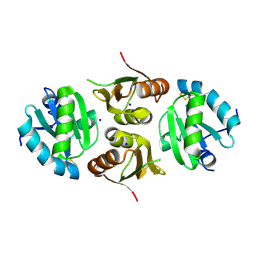 | | Structure of HuR RRM3 | | Descriptor: | SODIUM ION, Thioredoxin 1,ELAV-like protein 1 | | Authors: | Pabis, M, Sattler, M. | | Deposit date: | 2018-04-21 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs.
Nucleic Acids Res., 47, 2019
|
|
5EL3
 
 | | Structure of the KH domain of T-STAR | | Descriptor: | KH domain-containing, RNA-binding, signal transduction-associated protein 3, ... | | Authors: | Dominguez, C, Feracci, M. | | Deposit date: | 2015-11-04 | | Release date: | 2016-01-13 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural basis of RNA recognition and dimerization by the STAR proteins T-STAR and Sam68.
Nat Commun, 7, 2016
|
|
5ELT
 
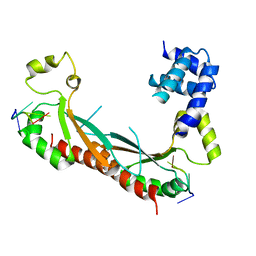 | |
