4S0W
 
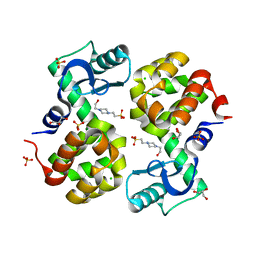 | | Wild type T4 lysozyme structure | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Snell, E.H, Snell, M.E. | | Deposit date: | 2015-01-07 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | Wild type T4 lysozyme structure
To be Published
|
|
6P7J
 
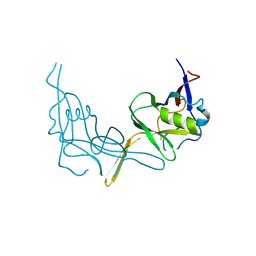 | |
6VE1
 
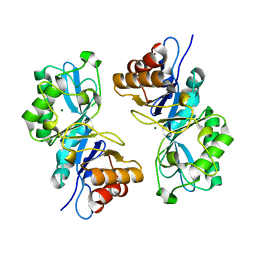 | |
8FUH
 
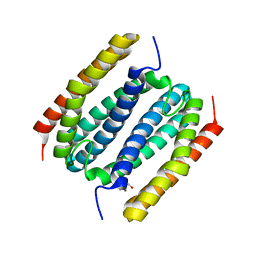 | |
8FVV
 
 | |
8FXD
 
 | |
6OBY
 
 | |
6NLR
 
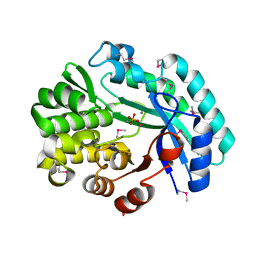 | | Crystal structure of the putative histidinol phosphatase hisK from Listeria monocytogenes with trinuclear metals determined by PIXE revealing sulphate ion in active site. Based on PIXE analysis and original date from 3DCP | | Descriptor: | CALCIUM ION, COBALT (II) ION, FE (III) ION, ... | | Authors: | Snell, E.H, Garman, E.F, Lowe, E.D. | | Deposit date: | 2019-01-09 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-Throughput PIXE as an Essential Quantitative Assay for Accurate Metalloprotein Structural Analysis: Development and Application.
J.Am.Chem.Soc., 142, 2020
|
|
6OE2
 
 | |
3TL4
 
 | |
8FW1
 
 | | Gluconobacter Ene-Reductase (GluER) mutant - PagER | | Descriptor: | FLAVIN MONONUCLEOTIDE, N-ethylmaleimide reductase | | Authors: | Dahagam, S, Page, C, Patterson, M.G, Hyster, T.K. | | Deposit date: | 2023-01-20 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Regioselective Radical Alkylation of Arenes Using Evolved Photoenzymes.
J.Am.Chem.Soc., 145, 2023
|
|
4H3S
 
 | | The Structure of Glutaminyl-tRNA Synthetase from Saccharomyces Cerevisiae | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, BROMIDE ION, ... | | Authors: | Snell, E.H, Grant, T.D. | | Deposit date: | 2012-09-14 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure of Yeast Glutaminyl-tRNA Synthetase and Modeling of Its Interaction with tRNA.
J.Mol.Biol., 425, 2013
|
|
