3RLE
 
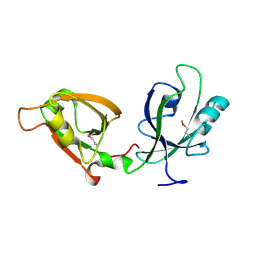 | | Crystal Structure of GRASP55 GRASP domain (residues 7-208) | | Descriptor: | Golgi reassembly-stacking protein 2 | | Authors: | Truschel, S.T, Sengupta, D, Foote, A, Heroux, A, Macbeth, M.R, Linstedt, A.D. | | Deposit date: | 2011-04-19 | | Release date: | 2011-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Structure of the Membrane-tethering GRASP Domain Reveals a Unique PDZ Ligand Interaction That Mediates Golgi Biogenesis.
J.Biol.Chem., 286, 2011
|
|
3DRI
 
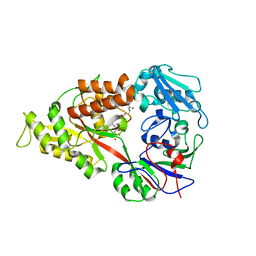 | | Crystal structure of Lactococcal OppA co-crystallized with an octamer peptide in an open conformation | | Descriptor: | Oligopeptide-binding protein oppA, peptide AASASA | | Authors: | Berntsson, R.P.-A, Doeven, M.K, Duurkens, R.H, Sengupta, D, Marrink, S.-J, Thunnissen, A.-M, Poolman, B, Slotboom, D.-J. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis for peptide selection by the transport receptor OppA
Embo J., 28, 2009
|
|
3DRJ
 
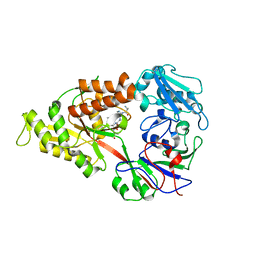 | | Crystal structure of Lactococcal OppA co-crystallized with pTH-related peptide in an open conformation | | Descriptor: | Oligopeptide-binding protein oppA, pTH-related peptide | | Authors: | Berntsson, R.P.-A, Doeven, M.K, Duurkens, R.H, Sengupta, D, Marrink, S.-J, Thunnissen, A.-M, Poolman, B, Slotboom, D.-J. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structural basis for peptide selection by the transport receptor OppA
Embo J., 28, 2009
|
|
3DRH
 
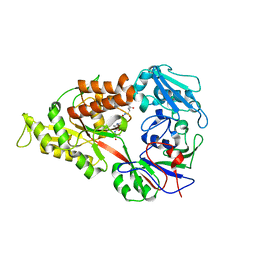 | | Crystal structure of Lactococcal OppA co-crystallized with Leu-enkephalin in an open conformation | | Descriptor: | Oligopeptide-binding protein oppA, peptide AAAAAA | | Authors: | Berntsson, R.P.-A, Doeven, M.K, Duurkens, R.H, Sengupta, D, Marrink, S.-J, Thunnissen, A.-M, Poolman, B, Slotboom, D.-J. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis for peptide selection by the transport receptor OppA
Embo J., 28, 2009
|
|
3DRF
 
 | | Lactococcal OppA complexed with an endogenous peptide in the closed conformation | | Descriptor: | Oligopeptide-binding protein oppA, endogenous peptide | | Authors: | Berntsson, R.P.-A, Doeven, M.K, Duurkens, R.H, Sengupta, D, Marrink, S.-J, Thunnissen, A.-M, Poolman, B, Slotboom, D.-J. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structural basis for peptide selection by the transport receptor OppA
Embo J., 28, 2009
|
|
3DRG
 
 | | Lactococcal OppA complexed with bradykinin in the closed conformation | | Descriptor: | Bradykinin, CHLORIDE ION, Oligopeptide-binding protein oppA | | Authors: | Berntsson, R.P.-A, Doeven, M.K, Duurkens, R.H, Sengupta, D, Marrink, S.-J, Thunnissen, A.-M, Poolman, B, Slotboom, D.-J. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural basis for peptide selection by the transport receptor OppA
Embo J., 28, 2009
|
|
3DRK
 
 | | Crystal structure of Lactococcal OppA co-crystallized with Neuropeptide S in an open conformation | | Descriptor: | Neuropeptide S, Oligopeptide-binding protein oppA | | Authors: | Berntsson, R.P.-A, Doeven, M.K, Duurkens, R.H, Sengupta, D, Marrink, S.-J, Thunnissen, A.-M, Poolman, B, Slotboom, D.-J. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis for peptide selection by the transport receptor OppA
Embo J., 28, 2009
|
|
5EMH
 
 | |
7CRQ
 
 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (2:1 binding mode) | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRR
 
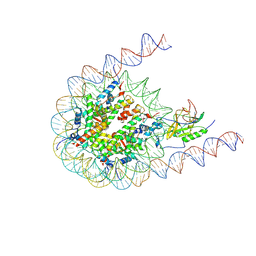 | | Native NSD3 bound to 187-bp nucleosome | | Descriptor: | DNA (168-MER), DNA(168-MER), Histone H2A, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRP
 
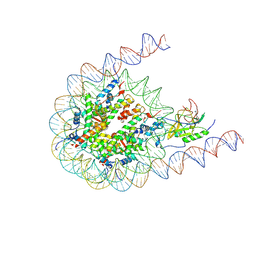 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (1:1 binding mode) | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRO
 
 | | NSD2 bearing E1099K/T1150A dual mutation in complex with 187-bp NCP | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
3T22
 
 | |
6K16
 
 | |
