1D8A
 
 | | E. COLI ENOYL REDUCTASE/NAD+/TRICLOSAN COMPLEX | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Levy, C.W, Roujeinikova, A, Sedelnikova, S, Baker, P.J, Stuitje, A.R, Slabas, A.R, Rice, D.W, Rafferty, J.B. | | Deposit date: | 1999-10-21 | | Release date: | 1999-10-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of triclosan activity.
Nature, 398, 1999
|
|
1D7O
 
 | | CRYSTAL STRUCTURE OF BRASSICA NAPUS ENOYL ACYL CARRIER PROTEIN REDUCTASE COMPLEXED WITH NAD AND TRICLOSAN | | Descriptor: | ENOYL-[ACYL-CARRIER PROTEIN] REDUCTASE (NADH) PRECURSOR, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Roujeinikova, A, Levy, C, Rowsell, S, Sedelnikova, S, Baker, P.J, Minshull, C.A, Mistry, A, Colls, J.G, Camble, R, Stuitje, A.R, Slabas, A.R, Rafferty, J.B, Pauptit, R.A, Viner, R, Rice, D.W. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic analysis of triclosan bound to enoyl reductase.
J.Mol.Biol., 294, 1999
|
|
1JPU
 
 | | Crystal Structure of Bacillus Stearothermophilus Glycerol Dehydrogenase | | Descriptor: | ZINC ION, glycerol dehydrogenase | | Authors: | Ruzheinikov, S.N, Burke, J, Sedelnikova, S, Baker, P.J, Taylor, R, Bullough, P.A, Muir, N.M, Gore, M.G, Rice, D.W. | | Deposit date: | 2001-08-03 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glycerol dehydrogenase. structure, specificity, and mechanism of a family III polyol dehydrogenase.
Structure, 9, 2001
|
|
1JQA
 
 | | Bacillus stearothermophilus glycerol dehydrogenase complex with glycerol | | Descriptor: | GLYCEROL, Glycerol Dehydrogenase, ZINC ION | | Authors: | Ruzheinikov, S.N, Burke, J, Sedelnikova, S, Baker, P.J, Taylor, R, Bullough, P.A, Muir, N.M, Gore, M.G, Rice, D.W. | | Deposit date: | 2001-08-04 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Glycerol dehydrogenase. structure, specificity, and mechanism of a family III polyol dehydrogenase.
Structure, 9, 2001
|
|
1LWJ
 
 | | CRYSTAL STRUCTURE OF T. MARITIMA 4-ALPHA-GLUCANOTRANSFERASE/ACARBOSE COMPLEX | | Descriptor: | 4-ALPHA-GLUCANOTRANSFERASE, CALCIUM ION, MODIFIED ACARBOSE PENTASACCHARIDE | | Authors: | Roujeinikova, A, Raasch, C, Sedelnikova, S, Liebl, W, Rice, D.W. | | Deposit date: | 2002-05-31 | | Release date: | 2002-08-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA 4-ALPHA-GLUCANOTRANSFERASE AND ITS ACARBOSE COMPLEX:
IMPLICATIONS FOR SUBSTRATE SPECIFICITY AND CATALYSIS
J.Mol.Biol., 321, 2002
|
|
1JQ5
 
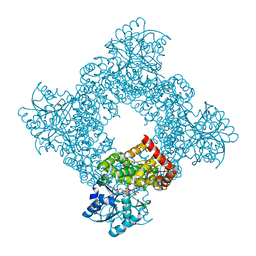 | | Bacillus Stearothermophilus Glycerol dehydrogenase complex with NAD+ | | Descriptor: | Glycerol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Ruzheinikov, S.N, Burke, J, Sedelnikova, S, Baker, P.J, Taylor, R, Bullough, P.A, Muir, N.M, Gore, M.G, Rice, D.W. | | Deposit date: | 2001-08-03 | | Release date: | 2001-10-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Glycerol dehydrogenase. structure, specificity, and mechanism of a family III polyol dehydrogenase.
Structure, 9, 2001
|
|
1LWH
 
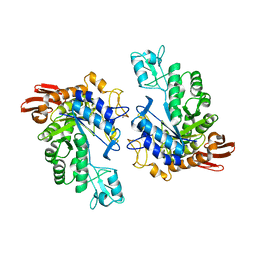 | | CRYSTAL STRUCTURE OF T. MARITIMA 4-ALPHA-GLUCANOTRANSFERASE | | Descriptor: | 4-alpha-glucanotransferase, CALCIUM ION | | Authors: | Roujeinikova, A, Raasch, C, Sedelnikova, S, Liebl, W, Rice, D.W. | | Deposit date: | 2002-05-31 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA 4-ALPHA-GLUCANOTRANSFERASE AND ITS ACARBOSE COMPLEX:
IMPLICATIONS FOR SUBSTRATE SPECIFICITY AND CATALYSIS
J.Mol.Biol., 321, 2002
|
|
1ZP7
 
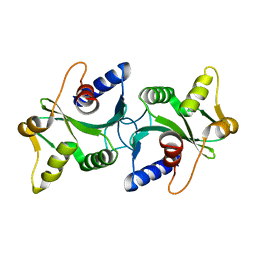 | | The structure of Bacillus subtilis RecU Holliday junction resolvase and its role in substrate selection and sequence specific cleavage. | | Descriptor: | Recombination protein U | | Authors: | McGregor, N, Ayora, S, Sedelnikova, S, Carrasco, B, Alonso, J.C, Thaw, P, Rafferty, J. | | Deposit date: | 2005-05-16 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure of Bacillus subtilis RecU Holliday Junction Resolvase and Its Role in Substrate Selection and Sequence-Specific Cleavage.
Structure, 13, 2005
|
|
3ZH9
 
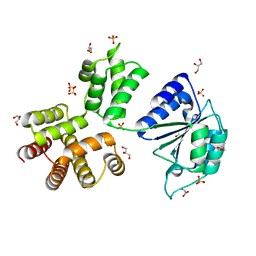 | | Bacillus subtilis DNA clamp loader delta protein (YqeN) | | Descriptor: | DELTA, GLYCEROL, SULFATE ION | | Authors: | Suwannachart, C, Sedelnikova, S, Soultanas, P, Oldham, N.J, Rafferty, J.B. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights Into the Structure and Assembly of the Bacillus Subtilis Clamp-Loader Complex and its Interaction with the Replicative Helicase.
Nucleic Acids Res., 41, 2013
|
|
1KKR
 
 | | CRYSTAL STRUCTURE OF CITROBACTER AMALONATICUS METHYLASPARTATE AMMONIA LYASE CONTAINING (2S,3S)-3-METHYLASPARTIC ACID | | Descriptor: | (2S,3S)-3-methyl-aspartic acid, 3-METHYLASPARTATE AMMONIA-LYASE, MAGNESIUM ION | | Authors: | Levy, C.W, Buckley, P.A, Sedelnikova, S, Kato, K, Asano, Y, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-12-10 | | Release date: | 2002-01-30 | | Last modified: | 2014-11-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into enzyme evolution revealed by the structure of methylaspartate ammonia lyase.
Structure, 10, 2002
|
|
1KKO
 
 | | CRYSTAL STRUCTURE OF CITROBACTER AMALONATICUS METHYLASPARTATE AMMONIA LYASE | | Descriptor: | 3-METHYLASPARTATE AMMONIA-LYASE, SULFATE ION | | Authors: | Levy, C.W, Buckley, P.A, Sedelnikova, S, Kato, Y, Asano, Y, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-12-10 | | Release date: | 2002-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Insights into enzyme evolution revealed by the structure of methylaspartate ammonia lyase.
Structure, 10, 2002
|
|
1SAY
 
 | | L-ALANINE DEHYDROGENASE COMPLEXED WITH PYRUVATE | | Descriptor: | L-ALANINE DEHYDROGENASE, PYRUVIC ACID | | Authors: | Baker, P.J, Sawa, Y, Shibata, H, Sedelnikova, S.E, Rice, D.W. | | Deposit date: | 1998-06-05 | | Release date: | 1999-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of the structure and substrate binding of Phormidium lapideum alanine dehydrogenase.
Nat.Struct.Biol., 5, 1998
|
|
1PJB
 
 | | L-ALANINE DEHYDROGENASE | | Descriptor: | L-ALANINE DEHYDROGENASE | | Authors: | Baker, P.J, Sawa, Y, Shibata, H, Sedelnikova, S.E, Rice, D.W. | | Deposit date: | 1998-06-05 | | Release date: | 1999-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of the structure and substrate binding of Phormidium lapideum alanine dehydrogenase.
Nat.Struct.Biol., 5, 1998
|
|
5FDK
 
 | |
2H8E
 
 | | Structure RusA D70N | | Descriptor: | Crossover junction endodeoxyribonuclease rusA | | Authors: | Macmaster, R.A. | | Deposit date: | 2006-06-07 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | RusA Holliday junction resolvase: DNA complex structure--insights into selectivity and specificity.
Nucleic Acids Res., 34, 2006
|
|
1CWU
 
 | | BRASSICA NAPUS ENOYL ACP REDUCTASE A138G MUTANT COMPLEXED WITH NAD+ AND THIENODIAZABORINE | | Descriptor: | 6-METHYL-2(PROPANE-1-SULFONYL)-2H-THIENO[3,2-D][1,2,3]DIAZABORININ-1-OL, ENOYL ACP REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Roujeinikova, A, Rafferty, J.B, Rice, D.W. | | Deposit date: | 1999-08-26 | | Release date: | 1999-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibitor binding studies on enoyl reductase reveal conformational changes related to substrate recognition.
J.Biol.Chem., 274, 1999
|
|
2H8C
 
 | | Structure of RusA D70N in complex with DNA | | Descriptor: | 5'-D(*CP*CP*GP*GP*TP*AP*CP*CP*GP*GP*T)-3', Crossover junction endodeoxyribonuclease rusA | | Authors: | Macmaster, R.A. | | Deposit date: | 2006-06-07 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | RusA Holliday junction resolvase: DNA complex structure--insights into selectivity and specificity.
Nucleic Acids Res., 34, 2006
|
|
4KTW
 
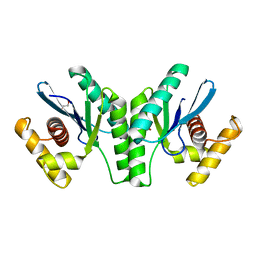 | | Lactococcus phage 67 RuvC - apo form | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, RuvC endonuclease | | Authors: | Rafferty, J.B, Green, V. | | Deposit date: | 2013-05-21 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Mutants of phage bIL67 RuvC with enhanced Holliday junction binding selectivity and resolution symmetry.
Mol.Microbiol., 89, 2013
|
|
4KTZ
 
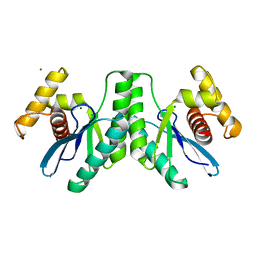 | | Lactococcus phage 67 RuvC | | Descriptor: | MAGNESIUM ION, RuvC endonuclease | | Authors: | Rafferty, J.B, Green, V. | | Deposit date: | 2013-05-21 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutants of phage bIL67 RuvC with enhanced Holliday junction binding selectivity and resolution symmetry.
Mol.Microbiol., 89, 2013
|
|
