4WEC
 
 | |
4XXV
 
 | |
4F4E
 
 | |
3KRE
 
 | |
3L0G
 
 | |
3KXQ
 
 | |
4F3P
 
 | |
3KZX
 
 | |
4TQT
 
 | |
4TRR
 
 | |
4WBS
 
 | |
4TV4
 
 | |
4WNY
 
 | |
4ZIY
 
 | |
5HGW
 
 | |
4HVT
 
 | |
5DLE
 
 | |
4HWG
 
 | |
4DDD
 
 | |
4ODJ
 
 | |
4EFF
 
 | |
4JWP
 
 | |
6VUD
 
 | |
4J07
 
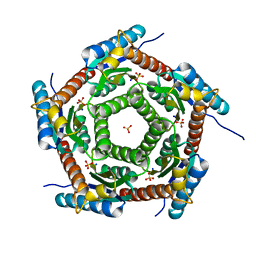 | | Crystal structure of a PROBABLE RIBOFLAVIN SYNTHASE, BETA CHAIN RIBH (6,7-dimethyl-8-ribityllumazine synthase, DMRL synthase, Lumazine synthase) from Mycobacterium leprae | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase, SODIUM ION, SULFATE ION | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2013-01-30 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a Probable Riboflavin Synthase, Beta chain RIBH (6,7-dimethyl-8-ribityllumazine synthase, DMRL synthase, Lumazine synthase) from Mycobacterium leprae
TO BE PUBLISHED
|
|
4K3Z
 
 | |
