1GW7
 
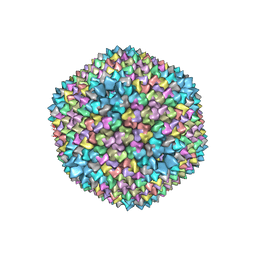 | | QUASI-ATOMIC RESOLUTION MODEL OF BACTERIOPHAGE PRD1 CAPSID, OBTAINED BY COMBINED CRYO-EM AND X-RAY CRYSTALLOGRAPHY. | | Descriptor: | MAJOR CAPSID PROTEIN | | Authors: | San Martin, C, Huiskonen, J, Bamford, J.K.H, Butcher, S.J, Fuller, S.D, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2002-03-08 | | Release date: | 2002-03-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Minor Proteins, Mobile Arms, and Membrane-Capsid Interactions in Bacteriophage Prd1 Capsid Assembly
Nat.Struct.Biol., 9, 2002
|
|
1GW8
 
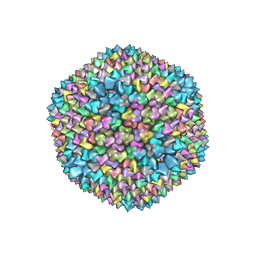 | | quasi-atomic resolution model of bacteriophage PRD1 sus607 mutant, obtained by combined cryo-EM and X-ray crystallography. | | Descriptor: | MAJOR CAPSID PROTEIN | | Authors: | San Martin, C, Huiskonen, J, Bamford, J.K.H, Butcher, S.J, Fuller, S.D, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2002-03-08 | | Release date: | 2002-03-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13.3 Å) | | Cite: | Minor Proteins, Mobile Arms and Membrane-Capsid Interactions in the Bacteriophage Prd1 Capsid.
Nat.Struct.Biol., 9, 2002
|
|
1HB9
 
 | | quasi-atomic resolution model of bacteriophage PRD1 wild type virion, obtained by combined cryo-EM and X-ray crystallography. | | Descriptor: | BACTERIOPHAGE PRD1 | | Authors: | San Martin, C, Burnett, R.M, De Haas, F, Heinkel, R, Rutten, T, Fuller, S.D, Butcher, S.J, Bamford, D.H. | | Deposit date: | 2001-04-13 | | Release date: | 2001-12-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Combined Em/X-Ray Imaging Yields a Quasi-Atomic Model of the Adenovirus-Related Bacteriophage Prd1 and Shows Key Capsid and Membrane Interactions
Structure, 9, 2001
|
|
6YBA
 
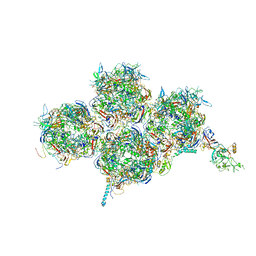 | | HAdV-F41 Capsid | | Descriptor: | Hexon protein, Hexon-interlacing protein, Penton protein, ... | | Authors: | Perez Illana, M, Martinez, M, Mangroo, C, Brown, M, Marabini, R, San Martin, C. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-10 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of enteric adenovirus HAdV-F41 highlights structural variations among human adenoviruses.
Sci Adv, 7, 2021
|
|
1W8X
 
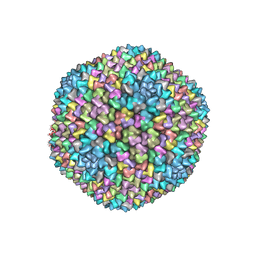 | | Structural analysis of PRD1 | | Descriptor: | MAJOR CAPSID PROTEIN (PROTEIN P3), PROTEIN P16, PROTEIN P30, ... | | Authors: | Abrescia, N.G.A, Cockburn, J.J.B, Grimes, J.M, Sutton, G.C, Diprose, J.M, Butcher, S.J, Fuller, S.D, San Martin, C, Burnett, R.M, Stuart, D.I, Bamford, D.H, Bamford, J.K.H. | | Deposit date: | 2004-10-01 | | Release date: | 2004-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Insights Into Assembly from Structural Analysis of Bacteriophage Prd1.
Nature, 432, 2004
|
|
1HB7
 
 | | quasi-atomic resolution model of bacteriophage PRD1 sus1 mutant, obtained by combined cryo-EM and X-ray crystallography. | | Descriptor: | BACTERIOPHAGE PRD1 SUS1 MUTANT CAPSID | | Authors: | San Martin, C, Burnett, R.M, De Haas, F, Heinkel, R, Rutten, T, Fuller, S.D, Butcher, S.J, Bamford, D.H. | | Deposit date: | 2001-04-12 | | Release date: | 2001-12-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Combined Em/X-Ray Imaging Yields a Quasi-Atomic Model of the Adenovirus-Related Bacteriophage Prd1 and Shows Key Capsid and Membrane Interactions.
Structure, 9, 2001
|
|
1HB5
 
 | | quasi-atomic resolution model of bacteriophage PRD1 P3-shell, obtained by combined cryo-EM and X-ray crystallography. | | Descriptor: | BACTERIOPHAGE PRD1 P3-SHELL | | Authors: | San Martin, C, Burnett, R.M, De Haas, F, Heinkel, R, Rutten, T, Fuller, S.D, Butcher, S.J, Bamford, D.H. | | Deposit date: | 2001-04-11 | | Release date: | 2001-12-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Combined Em/X-Ray Imaging Yields a Quasi-Atomic Model of the Adenovirus-Related Bacteriophage Prd1 and Shows Key Capsid and Membrane Interactions.
Structure, 9, 2001
|
|
4UMI
 
 | |
6QI5
 
 | | Near Atomic Structure of an Atadenovirus Shows a possible gene duplication event and Intergenera Variations in Cementing Proteins | | Descriptor: | Hexon protein, PIIIa, Penton protein, ... | | Authors: | Condezo, G.N, Marabini, R, Gomez-Blanco, J, SanMartin, C. | | Deposit date: | 2019-01-17 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Near-atomic structure of an atadenovirus reveals a conserved capsid-binding motif and intergenera variations in cementing proteins.
Sci Adv, 7, 2021
|
|
4EKF
 
 | |
7OOK
 
 | | Bacteriophage PRD1 Major Capsid Protein P3 in complex with CPZ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, CHLORIDE ION, ... | | Authors: | Duyvesteyn, H.M.E, Peccati, F, Martinez-Castillo, A, Jimenez-Oses, G, Oksanen, H.M, Stuart, D.I, Abrescia, N.G.A. | | Deposit date: | 2021-05-27 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Bacteriophage PRD1 as a nanoscaffold for drug loading
Nanoscale, 13, 2021
|
|
5G5O
 
 | | Structure of the snake adenovirus 1 hexon-interlacing LH3 protein, native | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nguyen, T.H, Singh, A.K, Albala-Perez, B, van Raaij, M.J. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Reptilian Adenovirus Reveals a Phage Tailspike Fold Stabilizing a Vertebrate Virus Capsid.
Structure, 25, 2017
|
|
5G5N
 
 | | Structure of the snake adenovirus 1 hexon-interlacing LH3 protein, methylmercury chloride derivative | | Descriptor: | CHLORIDE ION, GLYCEROL, LH3 HEXON-INTERLACING CAPSID PROTEIN, ... | | Authors: | Nguyen, T.H, Singh, A.K, Albala-Perez, B, van Raaij, M.J. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a Reptilian Adenovirus Reveals a Phage Tailspike Fold Stabilizing a Vertebrate Virus Capsid.
Structure, 25, 2017
|
|
4D1F
 
 | |
4D1G
 
 | | Crystal structure of the fiber head domain of the Atadenovirus snake adenovirus 1, native, second P212121 crystal form | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, FIBER PROTEIN, SULFATE ION | | Authors: | Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-05-01 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the fibre head domain of the Atadenovirus Snake Adenovirus 1.
PLoS ONE, 9, 2014
|
|
4D0U
 
 | | Crystal structure of the fiber head domain of the Atadenovirus snake adenovirus 1, selenomethionine-derivative | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, FIBER PROTEIN, SULFATE ION | | Authors: | Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-04-30 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the fibre head domain of the Atadenovirus Snake Adenovirus 1.
PLoS ONE, 9, 2014
|
|
4D0V
 
 | | Crystal structure of the fiber head domain of the Atadenovirus snake adenovirus 1, native, I213 crystal form | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, FIBER PROTEIN, SULFATE ION | | Authors: | Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-04-30 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the fibre head domain of the Atadenovirus Snake Adenovirus 1.
PLoS ONE, 9, 2014
|
|
8EQO
 
 | | Crystal structure of E.coli DsbA mutant K58A | | Descriptor: | COPPER (II) ION, GLYCEROL, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2022-10-08 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | A Buried Water Network Modulates the Activity of the Escherichia coli Disulphide Catalyst DsbA.
Antioxidants, 12, 2023
|
|
8EQQ
 
 | | Crystal structure of E.coli DsbA mutant E37A | | Descriptor: | CITRATE ANION, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A Buried Water Network Modulates the Activity of the Escherichia coli Disulphide Catalyst DsbA.
Antioxidants, 12, 2023
|
|
8EOC
 
 | | Crystal structure of E.coli DsbA mutant E24A/K58A | | Descriptor: | COPPER (II) ION, GLYCEROL, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2022-10-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A Buried Water Network Modulates the Activity of the Escherichia coli Disulphide Catalyst DsbA.
Antioxidants, 12, 2023
|
|
8EQR
 
 | | Crystal structure of E.coli DsbA mutant E24A | | Descriptor: | DI(HYDROXYETHYL)ETHER, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Buried Water Network Modulates the Activity of the Escherichia coli Disulphide Catalyst DsbA.
Antioxidants, 12, 2023
|
|
8EQP
 
 | | Crystal structure of E.coli DsbA mutant E24A/E37A/K58A | | Descriptor: | CITRATE ANION, GLYCEROL, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Buried Water Network Modulates the Activity of the Escherichia coli Disulphide Catalyst DsbA.
Antioxidants, 12, 2023
|
|
