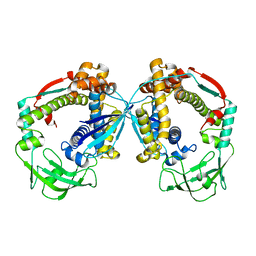
6D6S
 
 | |
2RPV
 
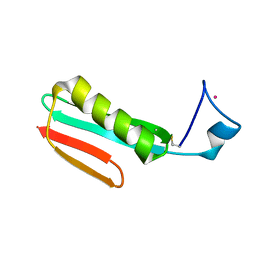 | | Solution Structure of GB1 with LBT probe | | Descriptor: | Immunoglobulin G-binding protein G, LANTHANUM (III) ION | | Authors: | Saio, T, Ogura, K, Yokochi, M, Kobashigawa, Y, Inagaki, F. | | Deposit date: | 2008-10-28 | | Release date: | 2009-09-15 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Two-point anchoring of a lanthanide-binding peptide to a target protein enhances the paramagnetic anisotropic effect
J.Biomol.Nmr, 44, 2009
|
|
2KTR
 
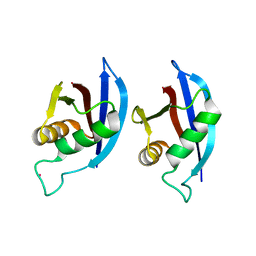 | | NMR structure of p62 PB1 dimer determined based on PCS | | Descriptor: | Sequestosome-1, TERBIUM(III) ION | | Authors: | Saio, T, Yokochi, M, Kumeta, H, Inagaki, F. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | PCS-based structure determination of protein-protein complexes
J.Biomol.Nmr, 46, 2010
|
|
2JNV
 
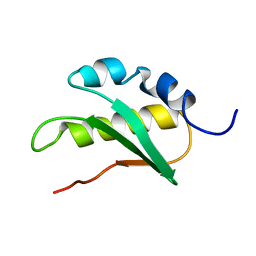 | | Solution structure of C-terminal domain of NifU-like protein from Oryza sativa | | Descriptor: | NifU-like protein 1, chloroplast | | Authors: | Saio, T, Ogura, K, Kumeta, H, Yokochi, M, Katoh, S, Katoh, E, Inagaki, F, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The cooperative role of OsCnfU-1A domain I and domain II in the iron sulphur cluster transfer process as revealed by NMR
J.Biochem.(Tokyo), 142, 2007
|
|
2MLX
 
 | | NMR structure of E. coli Trigger Factor in complex with unfolded PhoA220-310 | | Descriptor: | Alkaline phosphatase, Trigger factor | | Authors: | Saio, T, Guan, X, Rossi, P, Economou, A, Kalodimos, C.G. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for protein antiaggregation activity of the trigger factor chaperone.
Science, 344, 2014
|
|
2MLY
 
 | | NMR structure of E. coli Trigger Factor in complex with unfolded PhoA1-150 | | Descriptor: | Alkaline phosphatase, Trigger factor | | Authors: | Saio, T, Guan, X, Rossi, P, Economou, A, Kalodimos, C.G. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for protein antiaggregation activity of the trigger factor chaperone.
Science, 344, 2014
|
|
2MLZ
 
 | | NMR structure of E. coli Trigger Factor in complex with unfolded PhoA365-471 | | Descriptor: | Alkaline phosphatase, Trigger factor | | Authors: | Saio, T, Guan, X, Rossi, P, Economou, A, Kalodimos, C.G. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for protein antiaggregation activity of the trigger factor chaperone.
Science, 344, 2014
|
|
5ZR0
 
 | | Solution structure of peptidyl-prolyl cis/trans isomerase domain of Trigger Factor in complex with MBP | | Descriptor: | Maltose-binding periplasmic protein,Trigger factor | | Authors: | Kawagoe, S, Nakagawa, H, Kumeta, H, Ishimori, K, Saio, T. | | Deposit date: | 2018-04-21 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into prolinecis/transisomerization of unfolded proteins catalyzed by the trigger factor chaperone.
J. Biol. Chem., 293, 2018
|
|
2RSE
 
 | | NMR structure of FKBP12-mTOR FRB domain-rapamycin complex structure determined based on PCS | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Serine/threonine-protein kinase mTOR, TERBIUM(III) ION | | Authors: | Kobashigawa, Y, Ushio, M, Saio, T, Inagaki, F. | | Deposit date: | 2012-01-25 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Convenient method for resolving degeneracies due to symmetry of the magnetic susceptibility tensor and its application to pseudo contact shift-based protein-protein complex structure determination.
J.Biomol.Nmr, 53, 2012
|
|
5JTL
 
 | | The structure of chaperone SecB in complex with unstructured proPhoA | | Descriptor: | Alkaline phosphatase, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
5JTN
 
 | | The structure of chaperone SecB in complex with unstructured proPhoA binding site c | | Descriptor: | Alkaline phosphatase, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
5JTM
 
 | | The structure of chaperone SecB in complex with unstructured PhoA binding site a | | Descriptor: | Alkaline phosphatase, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
5JTQ
 
 | | The structure of chaperone SecB in complex with unstructured MBP binding site d | | Descriptor: | Maltose-binding periplasmic protein, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
5JTO
 
 | | The structure of chaperone SecB in complex with unstructured proPhoA binding site d | | Descriptor: | Alkaline phosphatase, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
5JTP
 
 | | The structure of chaperone SecB in complex with unstructured proPhoA binding site e | | Descriptor: | Alkaline phosphatase, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
5JTR
 
 | | The structure of chaperone SecB in complex with unstructured MBP binding site e | | Descriptor: | Maltose-binding periplasmic protein, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
2N9I
 
 | | Solution structure of reduced human cytochrome c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Imai, M, Saio, T, Kumeta, H, Uchida, T, Inagaki, F, Ishimori, K. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Investigation of the redox-dependent modulation of structure and dynamics in human cytochrome c
Biochem.Biophys.Res.Commun., 469, 2016
|
|
2N9J
 
 | | Solution structure of oxidized human cytochrome c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Imai, M, Saio, T, Kumeta, H, Uchida, T, Inagaki, F, Ishimori, K. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Investigation of the redox-dependent modulation of structure and dynamics in human cytochrome c.
Biochem.Biophys.Res.Commun., 469, 2016
|
|
2KKC
 
 | | NMR structure of the p62 PB1 domain | | Descriptor: | Sequestosome-1 | | Authors: | Yokochi, M, Inagaki, F. | | Deposit date: | 2009-06-18 | | Release date: | 2009-09-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the p62 PB1 domain, a key protein in autophagy and NF-kappaB signaling pathway
J.Biomol.Nmr, 45, 2009
|
|
