3L47
 
 | |
3L4A
 
 | |
3L4L
 
 | |
3QME
 
 | |
1RKB
 
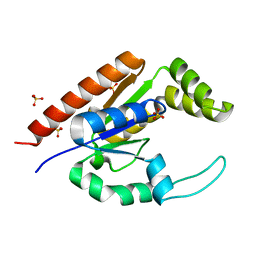 | | The structure of adrenal gland protein AD-004 | | Descriptor: | LITHIUM ION, Protein AD-004, SULFATE ION | | Authors: | Ren, H, Liang, Y, Bennett, M, Su, X.D. | | Deposit date: | 2003-11-21 | | Release date: | 2005-01-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human adenylate kinase 6: An adenylate kinase localized to the cell nucleus
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6YID
 
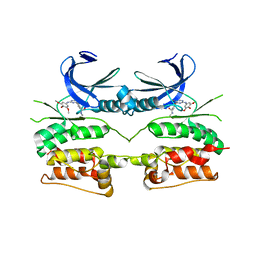 | | Crystal structure of ULK2 in complex with SBI-0206965 | | Descriptor: | 2-({5-bromo-2-[(3,4,5-trimethoxyphenyl)amino]pyrimidin-4-yl}oxy)-N-methylbenzene-1-carboximidic acid, Serine/threonine-protein kinase ULK2 | | Authors: | Chaikuad, A, Ren, H, Bakas, N.A, Lambert, L.J, Cosford, N.D.P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-01 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, Synthesis, and Characterization of an Orally Active Dual-Specific ULK1/2 Autophagy Inhibitor that Synergizes with the PARP Inhibitor Olaparib for the Treatment of Triple-Negative Breast Cancer.
J.Med.Chem., 63, 2020
|
|
3PJI
 
 | |
3BO5
 
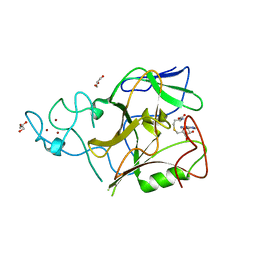 | | Crystal structure of methyltransferase domain of human Histone-lysine N-methyltransferase SETMAR | | Descriptor: | GLYCEROL, Histone-lysine N-methyltransferase SETMAR, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Lunin, V.V, Wu, H, Ren, H, Dobrovetsky, E, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The Crystal Structure of Methyltransferase Domain of Human Histone-lysine N-methyltransferase SETMAR in Complex With AdoHcy.
To be Published
|
|
5C20
 
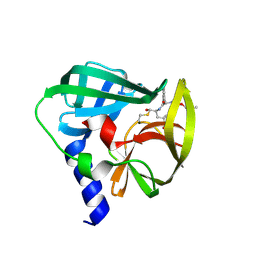 | | Crystal structure of EV71 3C Proteinase in complex with Compound 2 | | Descriptor: | 2-methylpropyl N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate, 3C proteinase | | Authors: | Zhang, L, Huang, G, Cai, Q, Zhao, C, Ren, H, Li, P, Li, N, Chen, S, Li, J, Lin, T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Optimize the interactions at S4 with efficient inhibitors targeting 3C proteinase from enterovirus 71
J.Mol.Recognit., 29, 2016
|
|
5C1U
 
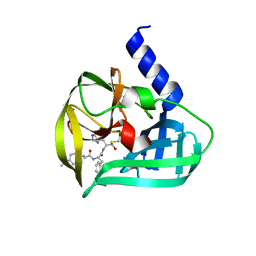 | | Crystal structure of EV71 3C Proteinase in complex with Compound Xb | | Descriptor: | (2S)-2-[[(E)-3-[4-(dimethylamino)phenyl]prop-2-enoyl]amino]-N-[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3-phenyl-propanamide, 3C proteinase | | Authors: | Zhang, L, Huang, G, Cai, Q, Zhao, C, Ren, H, Li, P, Li, N, Chen, S, Li, J, Lin, T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Optimize the interactions at S4 with efficient inhibitors targeting 3C proteinase from enterovirus 71
J.Mol.Recognit., 29, 2016
|
|
5C1X
 
 | | Crystal structure of EV71 3C Proteinase in complex with Compound VIII | | Descriptor: | (phenylmethyl) N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate, 3C proteinase | | Authors: | Zhang, L, Huang, G, Cai, Q, Zhao, C, Ren, H, Li, P, Li, N, Chen, S, Li, J, Lin, T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Optimize the interactions at S4 with efficient inhibitors targeting 3C proteinase from enterovirus 71
J.Mol.Recognit., 29, 2016
|
|
5C1Y
 
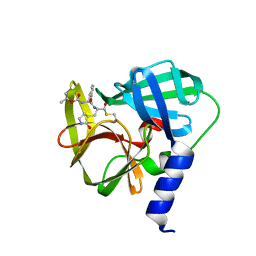 | | Crystal structure of EV71 3C Proteinase in complex with Compound 1 | | Descriptor: | 3C proteinase, propan-2-yl N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate | | Authors: | Zhang, L, Huang, G, Cai, Q, Zhao, C, Ren, H, Li, P, Li, N, Chen, S, Li, J, Lin, T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Optimize the interactions at S4 with efficient inhibitors targeting 3C proteinase from enterovirus 71
J.Mol.Recognit., 29, 2016
|
|
4BBD
 
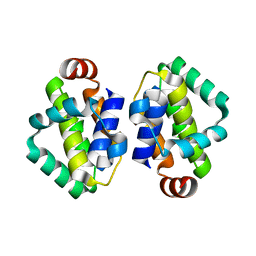 | | THE STRUCTURE OF VACCINIA VIRUS N1 R58Y MUTANT | | Descriptor: | N1L | | Authors: | Maluquer de Motes, C, Cooray, S, McGourty, K, Ren, H, Bahar, M.W, Stuart, D.I, Grimes, J.M, Graham, S.C, Smith, G.L. | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of Apoptosis and NF-kappaB Activation by Vaccinia Protein N1 Occur Via Distinct Binding Surfaces and Make Different Contributions to Virulence.
Plos Pathog., 7, 2011
|
|
4BBC
 
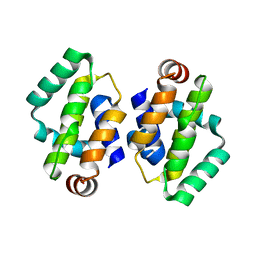 | | THE STRUCTURE OF VACCINIA VIRUS N1 R71Y MUTANT | | Descriptor: | N1L | | Authors: | Maluquer de Motes, C, Cooray, S, McGourty, K, Ren, H, Bahar, M.W, Stuart, D.I, Grimes, J.M, Graham, S.C, Smith, G.L. | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Inhibition of Apoptosis and NF-kappaB Activation by Vaccinia Protein N1 Occur Via Distinct Binding Surfaces and Make Different Contributions to Virulence.
Plos Pathog., 7, 2011
|
|
4BBB
 
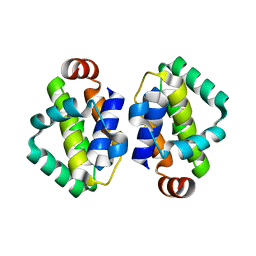 | | THE STRUCTURE OF VACCINIA VIRUS N1 Q61Y MUTANT | | Descriptor: | N1L | | Authors: | Maluquer de Motes, C, Cooray, S, McGourty, K, Ren, H, Bahar, M.W, Stuart, D.I, Grimes, J.M, Graham, S.C, Smith, G.L. | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Inhibition of Apoptosis and NF-kappaB Activation by Vaccinia Protein N1 Occur Via Distinct Binding Surfaces and Make Different Contributions to Virulence.
Plos Pathog., 7, 2011
|
|
2RCY
 
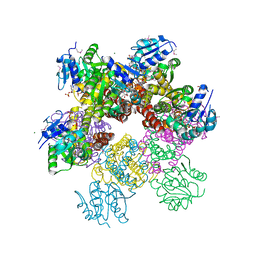 | | Crystal structure of Plasmodium falciparum pyrroline carboxylate reductase (MAL13P1.284) with NADP bound | | Descriptor: | GLYCEROL, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Wernimont, A.K, Lew, J, Lin, Y.H, Ren, H, Sun, X, Khuu, C, Hassanali, A, Wasney, G, Zhao, Y, Kozieradzki, I, Schapira, M, Bochkarev, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-20 | | Release date: | 2007-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Plasmodium falciparum pyrroline carboxylate reductase (MAL13P1.284) with NADP bound.
To be Published
|
|
3B7P
 
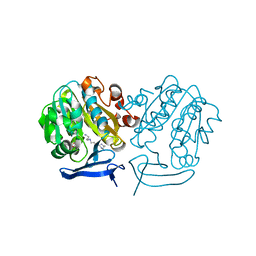 | | Crystal structure of spermidine synthase from Plasmodium falciparum in complex with spermine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, SPERMINE, Spermidine synthase | | Authors: | Qiu, W, Dong, A, Ren, H, Wu, H, Zhao, Y, Schapira, M, Wasney, G, Vedadi, M, Lew, J, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Plotnikov, A.N, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-31 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of spermidine synthase from Plasmodium falciparum in complex with spermine.
To be Published
|
|
1CFD
 
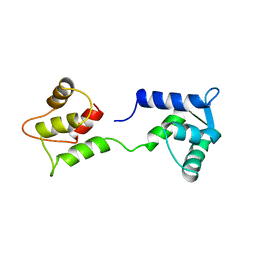 | | CALCIUM-FREE CALMODULIN | | Descriptor: | CALMODULIN | | Authors: | Kuboniwa, H, Tjandra, N, Grzesiek, S, Ren, H, Klee, C.B, Bax, A. | | Deposit date: | 1995-10-18 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calcium-free calmodulin.
Nat.Struct.Biol., 2, 1995
|
|
1CFC
 
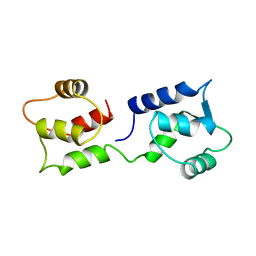 | | CALCIUM-FREE CALMODULIN | | Descriptor: | CALMODULIN | | Authors: | Kuboniwa, H, Tjandra, N, Grzesiek, S, Ren, H, Klee, C.B, Bax, A. | | Deposit date: | 1995-08-02 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calcium-free calmodulin.
Nat.Struct.Biol., 2, 1995
|
|
2I81
 
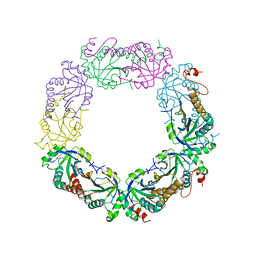 | | Crystal Structure of Plasmodium vivax 2-Cys Peroxiredoxin, Reduced | | Descriptor: | 2-Cys Peroxiredoxin | | Authors: | Artz, J.D, Qiu, W, Dong, A, Lew, J, Ren, H, Zhao, Y, Kozieradski, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-31 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Plasmodium vivax 2-Cys Peroxiredoxin, Reduced
To be published
|
|
2PWP
 
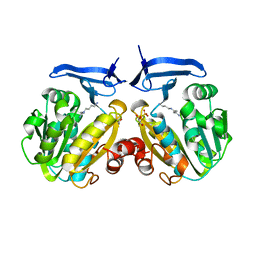 | | Crystal structure of spermidine synthase from Plasmodium falciparum in complex with spermidine | | Descriptor: | GLYCEROL, SPERMIDINE, SULFATE ION, ... | | Authors: | Qiu, W, Dong, A, Ren, H, Wu, H, Zhao, Y, Schapira, M, Wasney, G, Vedadi, M, Lew, J, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Plotnikov, A.N, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-11 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of spermidine synthase from Plasmodium falciparum in complex with spermidine.
To be Published
|
|
8C5N
 
 | | Sub-atomic resolution structure of the chitin-binding protein D (CbpD) from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, Chitin-binding protein CbpD | | Authors: | Cordara, G, Krengel, U, Golten, O, Vaaje-Kolstad, G, Vinther Soerensen, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.75 Å) | | Cite: | Immunization with lytic polysaccharide monooxygenase CbpD induces protective immunity against Pseudomonas aeruginosa pneumonia.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6XFT
 
 | |
8OHZ
 
 | | Yeast 20S proteasome in complex with a photoswitchable cepafungin derivative (transCep1) | | Descriptor: | (2~{S},3~{R})-2-[2-[4-[2-(4-ethylphenyl)hydrazinyl]phenyl]ethanoylamino]-~{N}-[(5~{S},8~{S},10~{S})-5-methyl-10-oxidanyl-2,7-bis(oxidanylidene)-1,6-diazacyclododec-8-yl]-3-oxidanyl-butanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Morstein, J, Amatuni, A, Schuster, A, Kuttenlochner, W, Ko, T, Groll, M, Adibekian, A, Renata, H, Trauner, D.H. | | Deposit date: | 2023-03-21 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Optical Control of Proteasomal Protein Degradation with a Photoswitchable Lipopeptide.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8OI1
 
 | | Yeast 20S proteasome in complex with a photoswitchable cepafungin derivative (transCep4) | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Morstein, J, Amatuni, A, Schuster, A, Kuttenlochner, W, Ko, T, Groll, M, Adibekian, A, Renata, H, Trauner, D.H. | | Deposit date: | 2023-03-21 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Optical Control of Proteasomal Protein Degradation with a Photoswitchable Lipopeptide.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
