5FA1
 
 | | The structure of the beta-3-deoxy-D-manno-oct-2-ulosonic acid transferase domain of WbbB | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Putative N-acetyl glucosaminyl transferase | | Authors: | Mallette, E, Ovchinnikova, O.G, Whitfield, C, Kimber, M.S. | | Deposit date: | 2015-12-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacterial beta-Kdo glycosyltransferases represent a new glycosyltransferase family (GT99).
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5FA0
 
 | | The structure of the beta-3-deoxy-D-manno-oct-2-ulosonic acid transferase domain from WbbB | | Descriptor: | CHLORIDE ION, Putative N-acetyl glucosaminyl transferase | | Authors: | Mallette, E, Ovchinnikova, O.G, Whitfield, C, Kimber, M.S. | | Deposit date: | 2015-12-10 | | Release date: | 2016-05-18 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bacterial beta-Kdo glycosyltransferases represent a new glycosyltransferase family (GT99).
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
6CI2
 
 | | Crystal structure of the formyltransferase PseJ from Anoxybacillus kamchatkensis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, formyltransferase PseJ | | Authors: | Reimer, J.M, Jiang, J, Harb, I, Schmeing, T.M. | | Deposit date: | 2018-02-23 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Insight into a Novel Formyltransferase and Evolution to a Nonribosomal Peptide Synthetase Tailoring Domain.
ACS Chem. Biol., 13, 2018
|
|
8QOY
 
 | | Capsular polysaccharide synthesis multienzyme of Actinobacillus Pleuropneumoniae serotype 3 | | Descriptor: | SULFATE ION, TagF-like capsule polymerase Cps3D, ZINC ION | | Authors: | Di Domenico, V, Litschko, C, Schulze, J, Bethe, A, Cifuente, J.O, Marina, A, Budde, I, Mast, T.A, Sulewska, M, Berger, M, Buettner, F, Gerardy-Schahn, R, Fiebig, T, Guerin, M.E. | | Deposit date: | 2023-09-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Transition transferases prime bacterial capsule polymerization.
Nat.Chem.Biol., 2024
|
|
6EDK
 
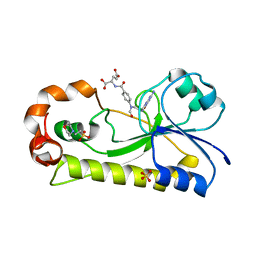 | | Crystal structure of the formyltransferase PseJ from Anoxybacillus kamchatkensis with N10-formyltetrahydrofolate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Formyltransferase PseJ, N-{4-[{[(6S)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}(formyl)amino]benzoyl}-L-glutamic acid, ... | | Authors: | Reimer, J.M, Harb, I, Schmeing, T.M. | | Deposit date: | 2018-08-09 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insight into a Novel Formyltransferase and Evolution to a Nonribosomal Peptide Synthetase Tailoring Domain.
ACS Chem. Biol., 13, 2018
|
|
8FUW
 
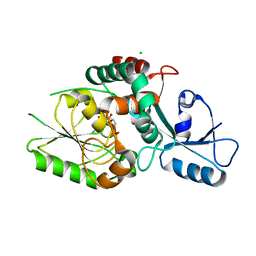 | | KpsC D160N Kdo adduct | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Kimber, M.S, Doyle, L, Whitfield, C. | | Deposit date: | 2023-01-18 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism and linkage specificities of the dual retaining beta-Kdo glycosyltransferase modules of KpsC from bacterial capsule biosynthesis.
J.Biol.Chem., 299, 2023
|
|
8FUX
 
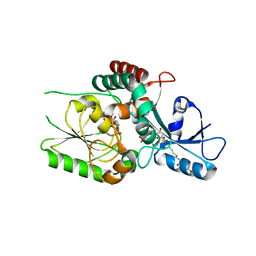 | | KpsC D160C ternary complex | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, 3-deoxy-beta-D-manno-oct-2-ulopyranosonic acid, CHLORIDE ION, ... | | Authors: | Kimber, M.S, Doyle, L, Whitfield, C. | | Deposit date: | 2023-01-18 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mechanism and linkage specificities of the dual retaining beta-Kdo glycosyltransferase modules of KpsC from bacterial capsule biosynthesis.
J.Biol.Chem., 299, 2023
|
|
6CI5
 
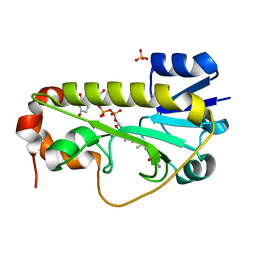 | | Crystal structure of the formyltransferase PseJ from Anoxybacillus kamchatkensis in complex with UDP-4,6-dideoxy-4-formamido-L-AltNAc and tetrahydrofolate | | Descriptor: | (2R,3R,4S,5R,6S)-3-(acetylamino)-5-(formylamino)-4-hydroxy-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), N-[4-({[(6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, SULFATE ION, ... | | Authors: | Reimer, J.M, Harb, I, Schmeing, T.M. | | Deposit date: | 2018-02-23 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.00003052 Å) | | Cite: | Structural Insight into a Novel Formyltransferase and Evolution to a Nonribosomal Peptide Synthetase Tailoring Domain.
ACS Chem. Biol., 13, 2018
|
|
6CI4
 
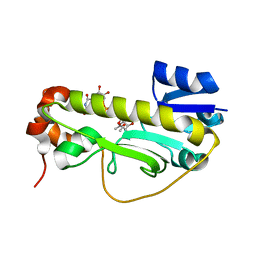 | | Crystal structure of the formyltransferase PseJ from Anoxybacillus kamchatkensis soaked with UDP-4-amino-4,6-dideoxy-L-AltNAc | | Descriptor: | (2R,3R,4S,5R,6S)-3-(acetylamino)-5-amino-4-hydroxy-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, formyltransferase PseJ | | Authors: | Harb, I, Reimer, J.M, Schmeing, T.M. | | Deposit date: | 2018-02-23 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.824068 Å) | | Cite: | Structural Insight into a Novel Formyltransferase and Evolution to a Nonribosomal Peptide Synthetase Tailoring Domain.
ACS Chem. Biol., 13, 2018
|
|
6U4B
 
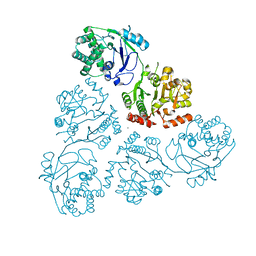 | | WbbM bifunctional glycosytransferase apo structure | | Descriptor: | MAGNESIUM ION, WbbM protein | | Authors: | Kimber, M.S, Mallette, E, Kamski-Hennekam, E.R, Gitalis, R. | | Deposit date: | 2019-08-25 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A bifunctional O-antigen polymerase structure reveals a new glycosyltransferase family.
Nat.Chem.Biol., 16, 2020
|
|
6MGB
 
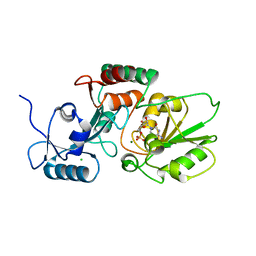 | | Thermosulfurimonas dismutans KpsC, beta Kdo 2,4 transferase | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, Capsular polysaccharide export system protein KpsC, ... | | Authors: | Doyle, L, Mallette, E, Kimber, M.S, Whitfield, C. | | Deposit date: | 2018-09-13 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biosynthesis of a conserved glycolipid anchor for Gram-negative bacterial capsules.
Nat.Chem.Biol., 15, 2019
|
|
6MGC
 
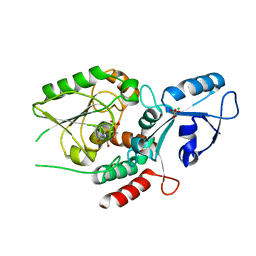 | | Escherichia coli KpsC, N-terminal domain | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, Capsule polysaccharide export protein KpsC, ... | | Authors: | Doyle, L, Mallette, E, Kimber, M.S. | | Deposit date: | 2018-09-13 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Biosynthesis of a conserved glycolipid anchor for Gram-negative bacterial capsules.
Nat.Chem.Biol., 15, 2019
|
|
6MGD
 
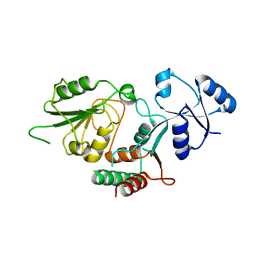 | | Thermosulfurimonas dismutans KpsC, beta Kdo 2,7 transferase | | Descriptor: | Capsular polysaccharide export system protein KpsC | | Authors: | Doyle, L, Mallette, E, Kimber, M.S, Whitfield, C. | | Deposit date: | 2018-09-13 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biosynthesis of a conserved glycolipid anchor for Gram-negative bacterial capsules.
Nat.Chem.Biol., 15, 2019
|
|
8CSE
 
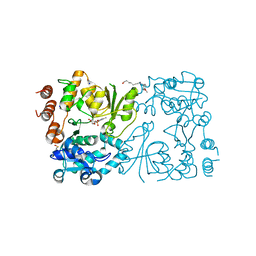 | | WbbB in complex with alpha-Rha-(1-3)-beta-GlcNAc acceptor | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, N-(8-hydroxyoctyl)-4-methoxybenzamide, N-acetyl glucosaminyl transferase, ... | | Authors: | Forrester, T.J.B, Kimber, M.S. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
8CSF
 
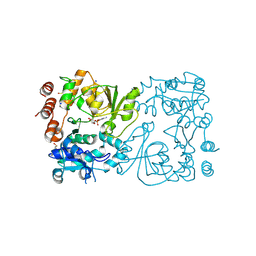 | | WbbB D232C-Kdo adduct + alpha-Rha(1,3)GlcNAc ternary complex | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, CYTIDINE-5'-MONOPHOSPHATE, N-acetyl glucosaminyl transferase, ... | | Authors: | Forrester, T.J.B, Kimber, M.S. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
8CSB
 
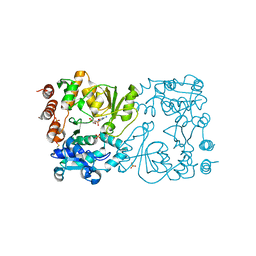 | | WbbB D232N in complex with CMP-beta-Kdo | | Descriptor: | CYTIDINE 5'-MONOPHOSPHATE 3-DEOXY-BETA-D-GULO-OCT-2-ULO-PYRANOSONIC ACID, CYTIDINE-5'-MONOPHOSPHATE, N-acetyl glucosaminyl transferase, ... | | Authors: | Forrester, T.J.B, Kimber, M.S. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
8CSD
 
 | | WbbB D232C Kdo adduct | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Forrester, T.J.B, Kimber, M.S. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
8CSC
 
 | | WbbB D232N-Kdo adduct | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Forrester, T.J.B, Kimber, M.S. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
