2J4M
 
 | | Double dockerin from Piromyces equi Cel45A | | Descriptor: | ENDOGLUCANASE 45A | | Authors: | Nagy, T, Tunnicliffe, R.B, Higgins, L.D, Walters, C, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2006-09-01 | | Release date: | 2007-09-25 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Characterization of a Double Dockerin from the Cellulosome of the Anaerobic Fungus Piromyces Equi.
J.Mol.Biol., 373, 2007
|
|
2J4N
 
 | | Double dockerin from Piromyces equi Cel45A | | Descriptor: | ENDOGLUCANASE 45A | | Authors: | Nagy, T, Tunnicliffe, R.B, Higgins, L.D, Walters, C, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2006-09-01 | | Release date: | 2007-09-25 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Characterization of a Double Dockerin from the Cellulosome of the Anaerobic Fungus Piromyces Equi.
J.Mol.Biol., 373, 2007
|
|
1H41
 
 | | Pseudomonas cellulosa E292A alpha-D-glucuronidase mutant complexed with aldotriuronic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-O-methyl-alpha-D-glucopyranuronic acid, ALPHA-GLUCURONIDASE, ... | | Authors: | Nurizzo, D, Nagy, T, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2002-09-25 | | Release date: | 2003-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Alpha-Glucuronidase,Glca67A,of Cellvibrio Japonicus Utilizes the Carboxylate and Methyl Groups of Aldobiouronic Acid as Important Substrate Recognition Determinants
J.Biol.Chem., 278, 2003
|
|
1GQI
 
 | | Structure of Pseudomonas cellulosa alpha-D-glucuronidase | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-GLUCURONIDASE, COBALT (II) ION, ... | | Authors: | Nurizzo, D, Nagy, T, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2001-11-26 | | Release date: | 2002-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The Structural Basis for Catalysis and Specificity of the Pseudomonas Cellulosa Alpha-Glucuronidase, Glca67A
Structure, 10, 2002
|
|
1GQK
 
 | | Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with glucuronic acid | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-D-GLUCURONIDASE, COBALT (II) ION, ... | | Authors: | Nurizzo, D, Nagy, T, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2001-11-26 | | Release date: | 2002-09-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structural Basis for Catalysis and Specificity of the Pseudomonas Cellulosa Alpha-Glucuronidase, Glca67A
Structure, 10, 2002
|
|
1GQJ
 
 | | Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with xylobiose | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-D-GLUCURONIDASE, COBALT (II) ION, ... | | Authors: | Nurizzo, D, Nagy, T, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2001-11-26 | | Release date: | 2002-09-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structural Basis for Catalysis and Specificity of the Pseudomonas Cellulosa Alpha-Glucuronidase, Glca67A
Structure, 10, 2002
|
|
1GQL
 
 | | Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with glucuronic acid and xylotriose | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-D-GLUCURONIDASE, COBALT (II) ION, ... | | Authors: | Nurizzo, D, Nagy, T, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2001-11-26 | | Release date: | 2002-09-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Structural Basis for Catalysis and Specificity of the Pseudomonas Cellulosa Alpha-Glucuronidase, Glca67A
Structure, 10, 2002
|
|
2VA0
 
 | | Differential regulation of the xylan degrading apparatus of Cellvibrio japonicus by a novel two component system | | Descriptor: | ABFS ARABINOFURANOSIDASE TWO COMPONENT SYSTEM SENSOR PROTEIN, CHLORIDE ION, PHOSPHATE ION | | Authors: | Murray, J.W, Emami, K, Topakas, E, Nagy, T, Henshaw, J, Jackson, K.A, Nelson, K.E, Mongodin, E.F, Lewis, R.J, Gilbert, H.J. | | Deposit date: | 2007-08-28 | | Release date: | 2008-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Regulation of the Xylan-Degrading Apparatus of Cellvibrio Japonicus by a Novel Two-Component System.
J.Biol.Chem., 284, 2009
|
|
7M0I
 
 | |
1UQZ
 
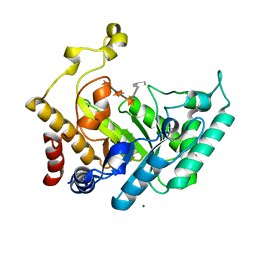 | | Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with 4-O-methyl glucuronic acid | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, CHLORIDE ION, ENDOXYLANASE, ... | | Authors: | Pell, G, Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-10-24 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Mechanisms by which Family 10 Glycoside Hydrolases Bind Decorated Substrates
J.Biol.Chem., 279, 2004
|
|
1UR1
 
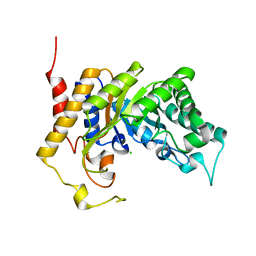 | | Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with arabinofuranose alpha-1,3 linked to xylobiose | | Descriptor: | CHLORIDE ION, ENDOXYLANASE, MAGNESIUM ION, ... | | Authors: | Pell, G, Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-10-24 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The Mechanisms by which Family 10 Glycoside Hydrolases Bind Decorated Substrates
J.Biol.Chem., 279, 2004
|
|
1UQY
 
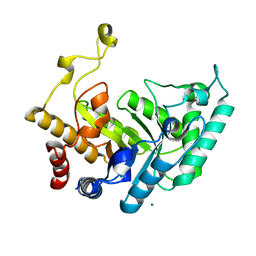 | | Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with xylopentaose | | Descriptor: | ENDOXYLANASE, MAGNESIUM ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose, ... | | Authors: | Pell, G, Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The Mechanisms by which Family 10 Glycoside Hydrolases Bind Decorated Substrates
J.Biol.Chem., 279, 2004
|
|
1UR2
 
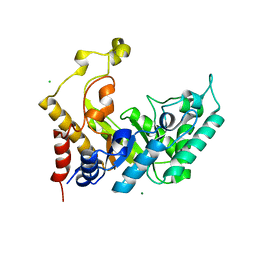 | | Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with arabinofuranose alpha 1,3 linked to xylotriose | | Descriptor: | CHLORIDE ION, ENDOXYLANASE, MAGNESIUM ION, ... | | Authors: | Pell, G, Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-10-24 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Mechanisms by which Family 10 Glycoside Hydrolases Bind Decorated Substrates
J.Biol.Chem., 279, 2004
|
|
2CCL
 
 | | THE S45A, T46A MUTANT OF THE TYPE I COHESIN-DOCKERIN COMPLEX FROM THE CELLULOSOME OF CLOSTRIDIUM THERMOCELLUM | | Descriptor: | CALCIUM ION, CELLULOSOMAL SCAFFOLDING PROTEIN A, ENDO-1,4-BETA-XYLANASE Y, ... | | Authors: | Carvalho, A.L, Dias, F.M.V, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Davies, G.J, Romao, M.J, Fontes, C.M.G.A. | | Deposit date: | 2006-01-16 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Evidence for a Dual Binding Mode of Dockerin Modules to Cohesins.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1E8R
 
 | | SOLUTION STRUCTURE OF TYPE X CBD | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Raghothama, S, Simpson, P.J, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2000-09-28 | | Release date: | 2000-10-03 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Cbm10 Cellulose Binding Module from Pseudomonas Xylanase A
Biochemistry, 39, 2000
|
|
1E8P
 
 | | Characterisation of the cellulose docking domain from Piromyces equi | | Descriptor: | Endoglucanase 45A | | Authors: | Raghothama, S, Eberhardt, R.Y, White, P, Hazlewood, G.P, Gilbert, H.J, Simpson, P.J, Williamson, M.P. | | Deposit date: | 2000-09-28 | | Release date: | 2001-09-07 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Characterization of a cellulosome dockerin domain from the anaerobic fungus Piromyces equi.
Nat. Struct. Biol., 8, 2001
|
|
1E8Q
 
 | | Characterisation of the cellulose docking domain from Piromyces equi | | Descriptor: | Endoglucanase 45A | | Authors: | Raghothama, S, Eberhardt, R.Y, White, P, Hazlewood, G.P, Gilbert, H.J, Simpson, P.J, Williamson, M.P. | | Deposit date: | 2000-09-28 | | Release date: | 2001-09-07 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Characterization of a cellulosome dockerin domain from the anaerobic fungus Piromyces equi.
Nat. Struct. Biol., 8, 2001
|
|
1QLD
 
 | |
