5I9K
 
 | | The structure of microsomal glutathione transferase 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GLUTATHIONE, Microsomal glutathione S-transferase 1, ... | | Authors: | Kuang, Q, Purhonen, P, Jegerschold, C, Morgenstern, R, Hebert, H. | | Deposit date: | 2016-02-20 | | Release date: | 2017-07-12 | | Last modified: | 2017-08-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Dead-end complex, lipid interactions and catalytic mechanism of microsomal glutathione transferase 1, an electron crystallography and mutagenesis investigation.
Sci Rep, 7, 2017
|
|
5IA9
 
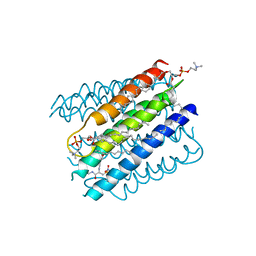 | | The structure of microsomal glutathione transferase 1 in complex with Meisenheimer complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-(S-GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXA-2,5-DIENE, Microsomal glutathione S-transferase 1, ... | | Authors: | Kuang, Q, Purhonen, P, Jegerschold, C, Morgenstern, R, Hebert, H. | | Deposit date: | 2016-02-21 | | Release date: | 2017-07-12 | | Last modified: | 2017-08-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Dead-end complex, lipid interactions and catalytic mechanism of microsomal glutathione transferase 1, an electron crystallography and mutagenesis investigation.
Sci Rep, 7, 2017
|
|
3HKK
 
 | |
3LEO
 
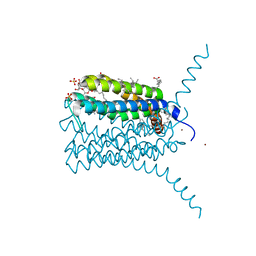 | | Structure of human Leukotriene C4 synthase mutant R31Q in complex with glutathione | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, GLUTATHIONE, GLYCEROL, ... | | Authors: | Niegowski, D, Martinez-Molina, D, Rinaldo-Matthis, A, Nordlund, P, Haeggstrom, J. | | Deposit date: | 2010-01-15 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Arginine 104 is a key catalytic residue in leukotriene C4 synthase.
J.Biol.Chem., 285, 2010
|
|
2H8A
 
 | |
3DWW
 
 | |
