1QWV
 
 | |
5BN5
 
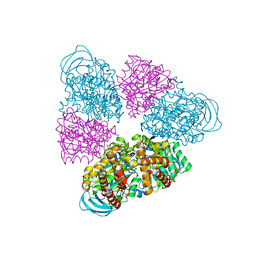 | | Structural basis for a unique ATP synthase core complex from Nanoarcheaum equitans | | Descriptor: | NEQ263, SULFATE ION, V-type ATP synthase alpha chain | | Authors: | Mohanty, S, Jobichen, C, Chichili, V.P.R, Sivaraman, J. | | Deposit date: | 2015-05-25 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Structural Basis for a Unique ATP Synthase Core Complex from Nanoarcheaum equitans
J.Biol.Chem., 290, 2015
|
|
5BN4
 
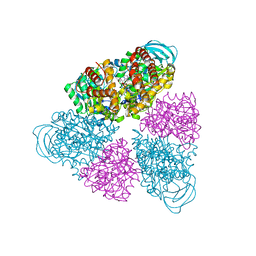 | | Structure of a unique ATP synthase NeqA-NeqB in complex with ANP from Nanoarcheaum equitans | | Descriptor: | MAGNESIUM ION, NEQ263, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Mohanty, S, Jobichen, C, Chichili, V.P.R, Sivaraman, J. | | Deposit date: | 2015-05-25 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structural Basis for a Unique ATP Synthase Core Complex from Nanoarcheaum equitans
J.Biol.Chem., 290, 2015
|
|
5BN3
 
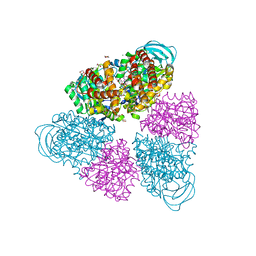 | | Structure of a unique ATP synthase NeqA-NeqB in complex with ADP from Nanoarcheaum equitans | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Mohanty, S, Jobichen, C, Chichili, V.P.R, Sivaraman, J. | | Deposit date: | 2015-05-25 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for a Unique ATP Synthase Core Complex from Nanoarcheaum equitans
J.Biol.Chem., 290, 2015
|
|
5BO5
 
 | | Structure of a unique ATP synthase subunit NeqB from Nanoarcheaum equitans | | Descriptor: | MAGNESIUM ION, NEQ263, SULFATE ION | | Authors: | Mohanty, S, Jobichen, C, Chichili, V.P.R, Sivaraman, J. | | Deposit date: | 2015-05-27 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Structural Basis for a Unique ATP Synthase Core Complex from Nanoarcheaum equitans
J.Biol.Chem., 290, 2015
|
|
1TWO
 
 | |
7UO6
 
 | |
1RKL
 
 | |
2JQU
 
 | |
2JQS
 
 | |
2L4T
 
 | | GIP/Glutaminase L peptide complex | | Descriptor: | Glutaminase L peptide, Tax1-binding protein 3 | | Authors: | Zoetewey, D.L, Ovee, M, Banerjee, M, Bhaskaran, R, Mohanty, S. | | Deposit date: | 2010-10-13 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Promiscuous binding at the crossroads of numerous cancer pathways: insight from the binding of glutaminase interacting protein with glutaminase L.
Biochemistry, 50, 2011
|
|
2L4S
 
 | | Promiscuous Binding at the Crossroads of Numerous Cancer Pathways: Insight from the Binding of GIP with Glutaminase L | | Descriptor: | Tax1-binding protein 3 | | Authors: | Zoetewey, D.L, Ovee, M, Banerjee, M, Bhaskaran, R, Mohanty, S. | | Deposit date: | 2010-10-13 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous binding at the crossroads of numerous cancer pathways: insight from the binding of glutaminase interacting protein with glutaminase L.
Biochemistry, 50, 2011
|
|
2LGZ
 
 | | Solution structure of STT3P | | Descriptor: | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3 | | Authors: | Huang, C, Bhaskaran, R, Mohanty, S. | | Deposit date: | 2011-08-03 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Eukaryotic N-Glycosylation Occurs via the Membrane-anchored C-terminal Domain of the Stt3p Subunit of Oligosaccharyltransferase.
J.Biol.Chem., 287, 2012
|
|
6ICE
 
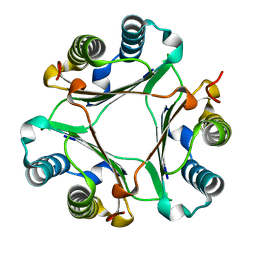 | | Crystal structure of Hamster MIF | | Descriptor: | Macrophage migration inhibitory factor | | Authors: | Sundaram, R, Vasudevan, D. | | Deposit date: | 2018-09-05 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Macrophage migration inhibitory factor of Syrian golden hamster shares structural and functional similarity with human counterpart and promotes pancreatic cancer.
Sci Rep, 9, 2019
|
|
6XCR
 
 | | NMR structure of Ost4 in DPC micelles | | Descriptor: | Oligosaccharyltransferase | | Authors: | Chaudhary, B.P. | | Deposit date: | 2020-06-09 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR and MD simulations reveal the impact of the V23D mutation on the function of yeast oligosaccharyltransferase subunit Ost4.
Glycobiology, 31, 2021
|
|
6XCU
 
 | |
