4FE1
 
 | | Improving the Accuracy of Macromolecular Structure Refinement at 7 A Resolution | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Fromme, R, Adams, P.D, Fromme, P, Levitt, M, Schroeder, G.F, Brunger, A.T. | | Deposit date: | 2012-05-29 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.9228 Å) | | Cite: | Improving the accuracy of macromolecular structure refinement at 7 A resolution.
Structure, 20, 2012
|
|
4V8R
 
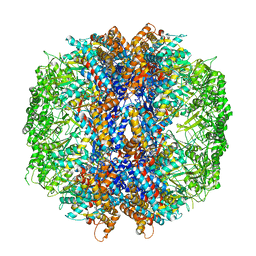 | | The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Kalisman, N, Schroder, G.F, Levitt, M. | | Deposit date: | 2012-03-28 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Crystal Structures of the Eukaryotic Chaperonin Cct Reveal its Functional Partitioning
Structure, 21, 2013
|
|
3TX8
 
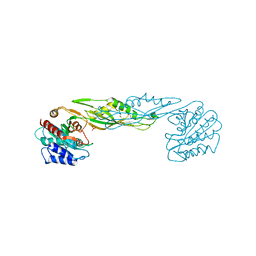 | | Crystal structure of a succinyl-diaminopimelate desuccinylase (ArgE) from Corynebacterium glutamicum ATCC 13032 at 2.97 A resolution | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Succinyl-diaminopimelate desuccinylase | | Authors: | Joint Center for Structural Genomics (JCSG), Brunger, A.T, Terwilliger, T.C, Read, R.J, Adams, P.D, Levitt, M, Schroder, G.F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.972 Å) | | Cite: | Application of DEN refinement and automated model building to a difficult case of molecular-replacement phasing: the structure of a putative succinyl-diaminopimelate desuccinylase from Corynebacterium glutamicum.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3LOS
 
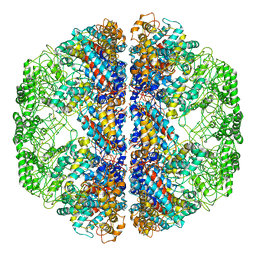 | | Atomic Model of Mm-cpn in the Closed State | | Descriptor: | Chaperonin | | Authors: | Zhang, J, Baker, M.L, Schroeder, G, Douglas, N.R, Reissmann, S, Jakana, J, Dougherty, M, Fu, C.J, Levitt, M, Ludtke, S.J, Frydman, J, Chiu, W. | | Deposit date: | 2010-02-04 | | Release date: | 2010-03-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mechanism of folding chamber closure in a group II chaperonin
Nature, 463, 2010
|
|
3J03
 
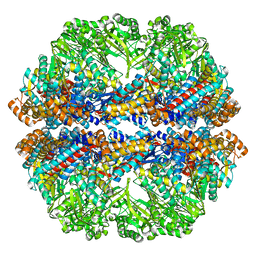 | | Lidless Mm-cpn in the closed state with ATP/AlFx | | Descriptor: | Lidless Mm-cpn | | Authors: | Zhang, J, Ma, B, DiMaio, F, Douglas, N.R, Joachimiak, L, Baker, D, Frydman, J, Levitt, M, Chiu, W. | | Deposit date: | 2011-02-10 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM structure of a group II chaperonin in the prehydrolysis ATP-bound state leading to lid closure.
Structure, 19, 2011
|
|
4B2T
 
 | | The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT ALPHA, T-COMPLEX PROTEIN 1 SUBUNIT BETA, T-COMPLEX PROTEIN 1 SUBUNIT DELTA, ... | | Authors: | Kalisman, N, Schroeder, G.F, Levitt, M. | | Deposit date: | 2012-07-17 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | The Crystal Structures of the Eukaryotic Chaperonin Cct Reveal its Functional Partitioning
Structure, 21, 2013
|
|
4A0O
 
 | | Symmetry-free cryo-EM map of TRiC in the nucleotide-free (apo) state | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-10 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.5 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
4A13
 
 | | model refined against symmetry-free cryo-EM map of TRiC-ADP | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11.3 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
4A0V
 
 | | model refined against the Symmetry-free cryo-EM map of TRiC-AMP-PNP | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.7 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
4A0W
 
 | | model built against symmetry-free cryo-EM map of TRiC-ADP-AlFx | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13.9 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
3IYF
 
 | | Atomic Model of the Lidless Mm-cpn in the Open State | | Descriptor: | Chaperonin | | Authors: | Zhang, J, Baker, M.L, Schroeder, G, Douglas, N.R, Reissmann, S, Jakana, J, Dougherty, M, Fu, C.J, Levitt, M, Ludtke, S.J, Frydman, J, Chiu, W. | | Deposit date: | 2009-10-23 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Mechanism of folding chamber closure in a group II chaperonin
Nature, 463, 2010
|
|
3GTK
 
 | | Backtracked RNA polymerase II complex with 18mer RNA | | Descriptor: | DNA (29-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
3GTO
 
 | | Backtracked RNA polymerase II complex with 15mer RNA | | Descriptor: | DNA (28-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
3GTG
 
 | | Backtracked RNA polymerase II complex with 12mer RNA | | Descriptor: | DNA (28-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.78 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
3GTJ
 
 | | Backtracked RNA polymerase II complex with 13mer RNA | | Descriptor: | DNA (28-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
3GTM
 
 | | Co-complex of Backtracked RNA polymerase II with TFIIS | | Descriptor: | DNA (28-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
3GTQ
 
 | | Backtracked RNA polymerase II complex induced by damage | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*CP*AP*TP*AP*AP*CP*CP*AP*CP*AP*GP*GP*CP*TP*CP*CP*TP*CP*TP*CP*CP*AP*TP*C)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
3GTL
 
 | | Backtracked RNA polymerase II complex with 13mer with G<>U mismatch | | Descriptor: | DNA (28-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
3GTP
 
 | | Backtracked RNA polymerase II complex with 24mer RNA | | Descriptor: | DNA (28-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
3J02
 
 | | Lidless D386A Mm-cpn in the pre-hydrolysis ATP-bound state | | Descriptor: | Lidless D386A Mm-cpn variant | | Authors: | Zhang, J, Ma, B, DiMaio, F, Douglas, N.R, Joachimiak, L, Baker, D, Frydman, J, Levitt, M, Chiu, W. | | Deposit date: | 2011-02-10 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Cryo-EM structure of a group II chaperonin in the prehydrolysis ATP-bound state leading to lid closure.
Structure, 19, 2011
|
|
6FGP
 
 | |
3ADK
 
 | |
4TNA
 
 | |
8LYZ
 
 | |
1EPY
 
 | | T4 LYSOZYME MUTANT, T21H/C54T/C97A/Q141H/T142H | | Descriptor: | CHLORIDE ION, COBALT (II) ION, LYSOZYME, ... | | Authors: | Wray, J.W, Baase, W.A, Ostheimer, G.J, Zhang, X.-J, Matthews, B.W. | | Deposit date: | 2000-03-29 | | Release date: | 2000-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Use of a non-rigid region in T4 lysozyme to design an adaptable metal-binding site.
Protein Eng., 13, 2000
|
|
