6H9O
 
 | |
6H9N
 
 | |
5FO3
 
 | | ZapC cell division regulator from E. coli | | Descriptor: | CELL DIVISION PROTEIN ZAPC | | Authors: | Ortiz, C, Kureisaite-Ciziene, D, Schmitz, F, Vicente, M, Lowe, J. | | Deposit date: | 2015-11-18 | | Release date: | 2015-11-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Z-Ring Associated Cell Division Protein Zapc from Escherichia Coli.
FEBS Lett., 589, 2015
|
|
4CJ7
 
 | | Structure of Crenactin, an archeal actin-like protein | | Descriptor: | ACTIN/ACTIN FAMILY PROTEIN, ADENOSINE-5'-DIPHOSPHATE | | Authors: | Izore, T, Duman, R.E, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crenactin from Pyrobaculum Calidifontis is Closely Related to Actin in Structure and Forms Steep Helical Filaments.
FEBS Lett., 588, 2014
|
|
7Q6D
 
 | | E. coli FtsA 1-405 ATP 3 Ni | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division protein FtsA, MAGNESIUM ION, ... | | Authors: | Nierhaus, T, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2021-11-06 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bacterial divisome protein FtsA forms curved antiparallel double filaments when binding to FtsN.
Nat Microbiol, 7, 2022
|
|
7Q6I
 
 | | Vibrio maritimus FtsA 1-396 ATP and FtsN 1-29, bent tetramers in double filament arrangement | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division protein FtsA, Cell division protein FtsN (polyAla model), ... | | Authors: | Nierhaus, T, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2021-11-07 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Bacterial divisome protein FtsA forms curved antiparallel double filaments when binding to FtsN.
Nat Microbiol, 7, 2022
|
|
7Q6G
 
 | | Xenorhabdus poinarii FtsA 1-396 ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division protein FtsA, MAGNESIUM ION | | Authors: | Nierhaus, T, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2021-11-07 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bacterial divisome protein FtsA forms curved antiparallel double filaments when binding to FtsN.
Nat Microbiol, 7, 2022
|
|
7Q6F
 
 | | Vibrio maritimus FtsA 1-396 ATP, double filament | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division protein FtsA, MAGNESIUM ION | | Authors: | Nierhaus, T, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2021-11-07 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Bacterial divisome protein FtsA forms curved antiparallel double filaments when binding to FtsN.
Nat Microbiol, 7, 2022
|
|
5MN5
 
 | | S. aureus FtsZ 12-316 T66W GTP Closed form (2TCm) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
5MN6
 
 | | S. aureus FtsZ 12-316 F138A GDP Closed form (3FCm) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
5N8P
 
 | |
5MN7
 
 | | S. aureus FtsZ 12-316 F138A GTP Closed form (3FCm) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
5MN4
 
 | | S. aureus FtsZ 12-316 F138A GDP Open form (1FOf) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
5MN8
 
 | | S. aureus FtsZ 12-316 F138A GTP Closed form (5FCm) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
6Z7P
 
 | | Composite model of the Caulobacter crescentus S-layer bound to the O-antigen of lipopolysaccharide | | Descriptor: | 4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose, CALCIUM ION, S-layer protein | | Authors: | Bharat, T.A.M, von Kugelgen, A. | | Deposit date: | 2020-06-01 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | In Situ Structure of an Intact Lipopolysaccharide-Bound Bacterial Surface Layer.
Cell, 180, 2020
|
|
8ONW
 
 | |
8QZO
 
 | |
7PEO
 
 | | Structure of the Caulobacter crescentus S-layer protein RsaA N-terminal domain bound to LPS and soaked with Holmium | | Descriptor: | 4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose, CALCIUM ION, HOLMIUM ATOM, ... | | Authors: | von Kugelgen, A, Bharat, T.A.M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.37 Å) | | Cite: | High-resolution mapping of metal ions reveals principles of surface layer assembly in Caulobacter crescentus cells.
Structure, 30, 2022
|
|
5LY3
 
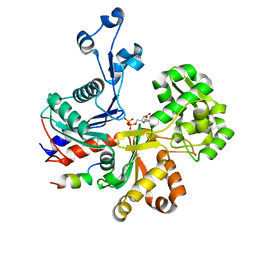 | |
6RIB
 
 | |
6RIA
 
 | |
6T72
 
 | | Structure of the RsaA N-terminal domain bound to LPS | | Descriptor: | 4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose, CALCIUM ION, S-layer protein | | Authors: | von Kuegelgen, A, Bharat, T.A.M. | | Deposit date: | 2019-10-21 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | In Situ Structure of an Intact Lipopolysaccharide-Bound Bacterial Surface Layer.
Cell, 180, 2020
|
|
5MW1
 
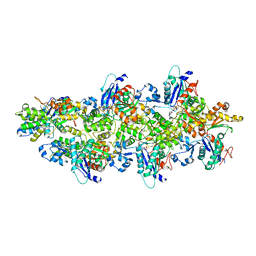 | |
5N97
 
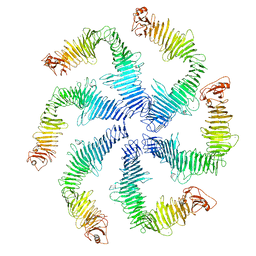 | | Structure of the C. crescentus S-layer | | Descriptor: | CALCIUM ION, S-layer protein rsaA | | Authors: | Bharat, T.A, Hagen, W.J, Briggs, J.A, Lowe, J. | | Deposit date: | 2017-02-24 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of the hexagonal surface layer on Caulobacter crescentus cells.
Nat Microbiol, 2, 2017
|
|
5O09
 
 | | BtubABC mini microtubule | | Descriptor: | Bacterial kinesin light chain, GUANOSINE-5'-DIPHOSPHATE, Tubulin, ... | | Authors: | Deng, X, Bharat, T.A.M, Lowe, J. | | Deposit date: | 2017-05-16 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Four-stranded mini microtubules formed by Prosthecobacter BtubAB show dynamic instability.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
