6Q4S
 
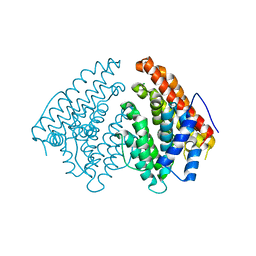 | | Crystal structure of a-eudesmol synthase | | Descriptor: | CHLORIDE ION, Pentalenene synthase | | Authors: | Correia Cordeiro, R.S, Hakansson, M, Logan, D.T, Kourist, R. | | Deposit date: | 2018-12-06 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of three novel sesquiterpene synthases from Streptomyces chartreusis NRRL 3882 and crystal structure of an alpha-eudesmol synthase.
J.Biotechnol., 297, 2019
|
|
6Y9K
 
 | |
6FTE
 
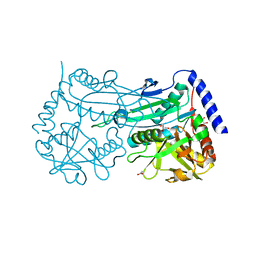 | | Crystal structure of an (R)-selective amine transaminase from Exophiala xenobiotica | | Descriptor: | ACETATE ION, Amine transaminase (fold IV), GLYCEROL, ... | | Authors: | Telzerow, A, Hakansson, M, Schurrmann, M, Schwab, H, Steiner, K. | | Deposit date: | 2018-02-21 | | Release date: | 2019-01-09 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Amine Transaminase from Exophiala xenobiotica - Crystal Structure and Engineering of a Fold IV Transaminase that Naturally Converts Biaryl Ketones
Acs Catalysis, 2018
|
|
7O6P
 
 | |
7O6Q
 
 | |
8A85
 
 | |
8ACS
 
 | | Crystal structure of FMO from Janthinobacterium svalbardensis | | Descriptor: | DI(HYDROXYETHYL)ETHER, FAD-dependent oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Polidori, N, Galuska, P, Gruber, K. | | Deposit date: | 2022-07-06 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Cold-Active Flavin-Dependent Monooxygenase from Janthinobacterium svalbardensis Unlocks Applications of Baeyer-Villiger Monooxygenases at Low Temperature.
Acs Catalysis, 13, 2023
|
|
6ZZ0
 
 | |
6ZZT
 
 | |
6ZYZ
 
 | | Structure of the borneol dehydrogenases of Salvia rosmarinus with NAD+ | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, Borneol dehydrogenase from salvia rosmarinus, CHLORIDE ION, ... | | Authors: | Dimos, N, Helmer, C.P.O, Loll, B. | | Deposit date: | 2020-08-03 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | A Structural View on the Stereospecificity of Plant Borneol-Type Dehydrogenases.
Chemcatchem, 13, 2021
|
|
8ADX
 
 | |
8B30
 
 | |
8C66
 
 | |
3ZPX
 
 | |
