2RFZ
 
 | | Crystal structure of cellobiohydrolase from Melanocarpus albomyces complexed with cellotriose | | Descriptor: | Cellulose 1,4-beta-cellobiosidase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Parkkinen, T, Koivula, A, Vehmaanper, J, Rouvinen, J. | | Deposit date: | 2007-10-02 | | Release date: | 2008-09-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of Melanocarpus albomyces cellobiohydrolase Cel7B in complex with cello-oligomers show high flexibility in the substrate binding
Protein Sci., 17, 2008
|
|
2RG0
 
 | | Crystal structure of cellobiohydrolase from Melanocarpus albomyces complexed with cellotetraose | | Descriptor: | Cellulose 1,4-beta-cellobiosidase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Parkkinen, T, Koivula, A, Vehmaanper, J, Rouvinen, J. | | Deposit date: | 2007-10-02 | | Release date: | 2008-09-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of Melanocarpus albomyces cellobiohydrolase Cel7B in complex with cello-oligomers show high flexibility in the substrate binding
Protein Sci., 17, 2008
|
|
2RFY
 
 | | Crystal structure of cellobiohydrolase from Melanocarpus albomyces complexed with cellobiose | | Descriptor: | Cellulose 1,4-beta-cellobiosidase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Parkkinen, T, Koivula, A, Vehmaanper, J, Rouvinen, J. | | Deposit date: | 2007-10-02 | | Release date: | 2008-09-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of Melanocarpus albomyces cellobiohydrolase Cel7B in complex with cello-oligomers show high flexibility in the substrate binding
Protein Sci., 17, 2008
|
|
2RFW
 
 | | Crystal Structure of Cellobiohydrolase from Melanocarpus albomyces | | Descriptor: | Cellulose 1,4-beta-cellobiosidase | | Authors: | Parkkinen, T, Koivula, A, Vehmaanper, J, Rouvinen, J. | | Deposit date: | 2007-10-02 | | Release date: | 2008-09-16 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of Melanocarpus albomyces cellobiohydrolase Cel7B in complex with cello-oligomers show high flexibility in the substrate binding
Protein Sci., 17, 2008
|
|
1GW0
 
 | | Crystal Structure of Laccase from Melanocarpus albomyces in Four Copper Form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hakulinen, N, Kiiskinen, L.-L, Kruus, K, Saloheimo, M, Koivula, A, Rouvinen, J. | | Deposit date: | 2002-03-01 | | Release date: | 2002-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a Laccase from Melanocarpus Albomyces with an Intact Trinuclear Copper Site
Nat.Struct.Biol., 9, 2002
|
|
2C1P
 
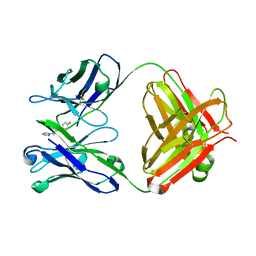 | | Fab-fragment of enantioselective antibody complexed with finrozole | | Descriptor: | 4-[(1S,2R)-3-(4-FLUOROPHENYL)-2-HYDROXY-1-(1H-1,2,4-TRIAZOL-1-YL)PROPYL]BENZONITRILE, IGH-4 PROTEIN, IGK-C PROTEIN | | Authors: | Parkkinen, T, Nevanen, T.K, Koivula, A, Rouvinen, J. | | Deposit date: | 2005-09-19 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of an Enantioselective Fab-Fragment in Free and Complex Forms.
J.Mol.Biol., 357, 2006
|
|
2C1O
 
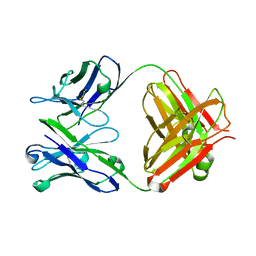 | | ENAIIHis Fab fragment in the free form | | Descriptor: | IGH-4 PROTEIN, IGK-C PROTEIN | | Authors: | Parkkinen, T, Nevanen, T.K, Koivula, A, Rouvinen, J. | | Deposit date: | 2005-09-19 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structures of an Enantioselective Fab-Fragment in Free and Complex Forms.
J.Mol.Biol., 357, 2006
|
|
1CB2
 
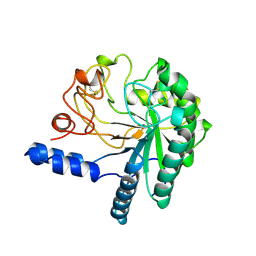 | | CELLOBIOHYDROLASE II, CATALYTIC DOMAIN, MUTANT Y169F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE II, alpha-D-mannopyranose | | Authors: | Kleywegt, G.J, Szardenings, M, Jones, T.A. | | Deposit date: | 1995-11-25 | | Release date: | 1996-10-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The active site of Trichoderma reesei cellobiohydrolase II: the role of tyrosine 169.
Protein Eng., 9, 1996
|
|
1HGW
 
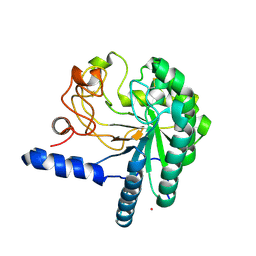 | | CEL6A D175A mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II), COBALT (II) ION, ... | | Authors: | Zou, J.-Y, Jones, T.A. | | Deposit date: | 2000-12-15 | | Release date: | 2002-01-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Active Site of Cellobiohydrolase Cel6A from Trichoderma Reesei: The Roles of Aspartic Acids D221 and D175
J.Am.Chem.Soc., 124, 2002
|
|
1HGY
 
 | | CEL6A D221A mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II), alpha-D-glucopyranose, ... | | Authors: | Zou, J.-Y, Kleywegt, G.J, Jones, T.A. | | Deposit date: | 2000-12-15 | | Release date: | 2002-01-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Active Site of Cellobiohydrolase Cel6A from Trichoderma Reesei: The Roles of Aspartic Acids D221 and D175
J.Am.Chem.Soc., 124, 2002
|
|
3CEL
 
 | | ACTIVE-SITE MUTANT E212Q DETERMINED AT PH 6.0 WITH CELLOBIOSE BOUND IN THE ACTIVE SITE | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1996-08-24 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activity studies and crystal structures of catalytically deficient mutants of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 264, 1996
|
|
4UR8
 
 | | Crystal structure of keto-deoxy-D-galactarate dehydratase complexed with 2-oxoadipic acid | | Descriptor: | 2-OXOADIPIC ACID, FORMIC ACID, KETO-DEOXY-D-GALACTARATE DEHYDRATASE | | Authors: | Taberman, H, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2014-06-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structure and Function of a Decarboxylating Agrobacterium Tumefaciens Keto-Deoxy-D-Galactarate Dehydratase.
Biochemistry, 53, 2014
|
|
4UR7
 
 | | Crystal structure of keto-deoxy-D-galactarate dehydratase complexed with pyruvate | | Descriptor: | FORMIC ACID, GLYCEROL, KETO-DEOXY-D-GALACTARATE DEHYDRATASE | | Authors: | Taberman, H, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2014-06-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structure and Function of a Decarboxylating Agrobacterium Tumefaciens Keto-Deoxy-D-Galactarate Dehydratase.
Biochemistry, 53, 2014
|
|
1EGN
 
 | | CELLOBIOHYDROLASE CEL7A (E223S, A224H, L225V, T226A, D262G) MUTANT | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE CEL7A, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION | | Authors: | Stahlberg, J, Harris, M, Jones, T.A. | | Deposit date: | 2000-02-16 | | Release date: | 2001-05-16 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering of a glycosidase Family 7 cellobiohydrolase to more alkaline pH optimum: the pH behaviour of Trichoderma reesei Cel7A and its E223S/ A224H/L225V/T226A/D262G mutant.
Biochem.J., 356, 2001
|
|
1Q2E
 
 | | CELLOBIOHYDROLASE CEL7A WITH LOOP DELETION 245-252 AND BOUND NON-HYDROLYSABLE CELLOTETRAOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, EXOCELLOBIOHYDROLASE I, ... | | Authors: | Stahlberg, J, Harris, M, Jones, T.A. | | Deposit date: | 2003-07-24 | | Release date: | 2003-11-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Cel7A.
A comparison with Phanerochaete chrysosporium Cel7D
J.Mol.Biol., 333, 2003
|
|
1Q2B
 
 | | CELLOBIOHYDROLASE CEL7A WITH DISULPHIDE BRIDGE ADDED ACROSS EXO-LOOP BY MUTATIONS D241C AND D249C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, EXOCELLOBIOHYDROLASE I | | Authors: | Stahlberg, J, Harris, M, Jones, T.A. | | Deposit date: | 2003-07-24 | | Release date: | 2003-11-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Cel7A.
A comparison with Phanerochaete chrysosporium Cel7D.
J.Mol.Biol., 333, 2003
|
|
5A03
 
 | | Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with xylose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ALDOSE-ALDOSE OXIDOREDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Taberman, H, Rouvinen, J, Parkkinen, T. | | Deposit date: | 2015-04-17 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Structure and Function of Caulobacter Crescentus Aldose-Aldose Oxidoreductase.
Biochem.J., 472, 2015
|
|
3PPS
 
 | | Crystal structure of an ascomycete fungal laccase from Thielavia arenaria | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Kallio, J.P, Rouvinen, J, Hakulinen, N. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an ascomycete fungal laccase from Thielavia arenaria--common structural features of asco-laccases.
Febs J., 278, 2011
|
|
4CEL
 
 | | ACTIVE-SITE MUTANT D214N DETERMINED AT PH 6.0 WITH NO LIGAND BOUND IN THE ACTIVE SITE | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1996-08-24 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activity studies and crystal structures of catalytically deficient mutants of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 264, 1996
|
|
2Q9O
 
 | | Near-atomic resolution structure of a Melanocarpus albomyces laccase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hakulinen, N, Rouvinen, J. | | Deposit date: | 2007-06-13 | | Release date: | 2008-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A near atomic resolution structure of a Melanocarpus albomyces laccase.
J.Struct.Biol., 162, 2008
|
|
5OYN
 
 | | Crystal structure of D-xylonate dehydratase in holo-form | | Descriptor: | Dehydratase, IlvD/Edd family, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rahman, M.M, Rouvinen, J, Hakulinen, N. | | Deposit date: | 2017-09-11 | | Release date: | 2018-01-24 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of D-xylonate dehydratase reveals functional features of enzymes from the Ilv/ED dehydratase family.
Sci Rep, 8, 2018
|
|
2IH8
 
 | | A low-dose crystal structure of a recombinant Melanocarpus albomyces laccase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hakulinen, N, Rouvinen, J. | | Deposit date: | 2006-09-26 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A crystallographic and spectroscopic study on the effect of X-ray radiation on the crystal structure of Melanocarpus albomyces laccase.
Biochem.Biophys.Res.Commun., 350, 2006
|
|
2IH9
 
 | | A high-dose crystal structure of a recombinant Melanocarbus albomyces laccase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hakulinen, N, Rouvinen, J. | | Deposit date: | 2006-09-26 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A crystallographic and spectroscopic study on the effect of X-ray radiation on the crystal structure of Melanocarpus albomyces laccase.
Biochem.Biophys.Res.Commun., 350, 2006
|
|
7CEL
 
 | | CBH1 (E217Q) IN COMPLEX WITH CELLOHEXAOSE AND CELLOBIOSE | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1997-09-24 | | Release date: | 1997-12-24 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 275, 1998
|
|
7PLD
 
 | |
