3KGG
 
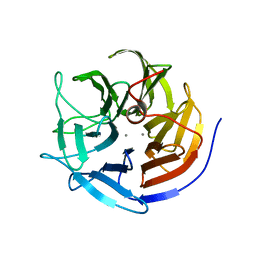 | | X-ray structure of perdeuterated diisopropyl fluorophosphatase (DFPase): Perdeuteration of proteins for neutron diffraction | | Descriptor: | CALCIUM ION, Diisopropyl-fluorophosphatase | | Authors: | Blum, M.-M, Tomanicek, S.J, John, H, Hanson, B.L, terjans, H.R, Schoenborn, B.P, Langan, P, Chen, J.C.-H. | | Deposit date: | 2009-10-29 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of perdeuterated diisopropyl fluorophosphatase (DFPase): perdeuteration of proteins for neutron diffraction.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2H9X
 
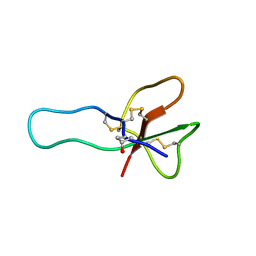 | | NMR structure for the CgNa toxin from the sea anemone Condylactis gigantea | | Descriptor: | Toxin CgNa | | Authors: | Lopez-Mendez, B, Perez-Castells, J, Gimenez-Gallego, G, Jimenez-Barbero, J. | | Deposit date: | 2006-06-12 | | Release date: | 2007-06-05 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | CgNa, a type I toxin from the giant Caribbean sea anemone Condylactis gigantea shows structural similarities to both type I and II toxins, as well as distinctive structural and functional properties(1).
Biochem.J., 406, 2007
|
|
8Q2D
 
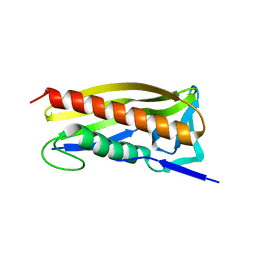 | |
8Q2C
 
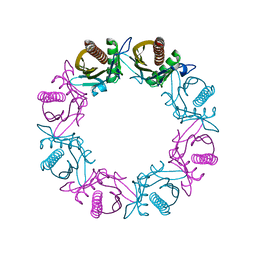 | |
4DH5
 
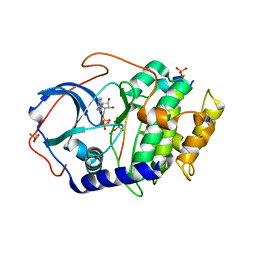 | | Room temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, ADP, Phosphate, and IP20 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DH1
 
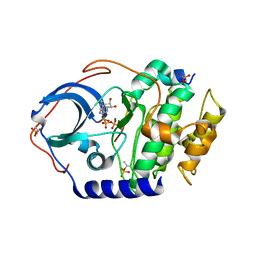 | | Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with low Mg2+, ATP and IP20 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DH7
 
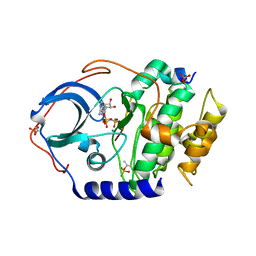 | | Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, AMP-PNP and IP20' | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DH3
 
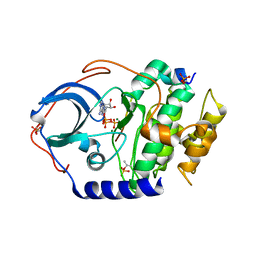 | | Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, ATP and IP20 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DH8
 
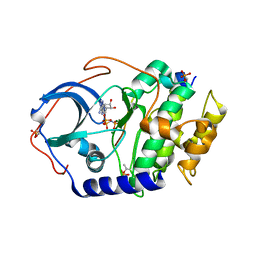 | | Room temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, AMP-PNP and IP20 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8OJG
 
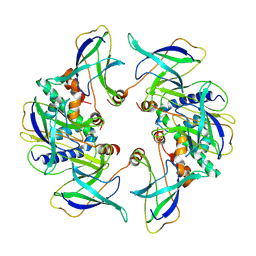 | | Structure of the MlaCD complex (2:6 stoichiometry) | | Descriptor: | Intermembrane phospholipid transport system binding protein MlaC, Intermembrane phospholipid transport system binding protein MlaD | | Authors: | Wotherspoon, P, Bui, S, Sridhar, P, Bergeron, J.R.C, Knowles, T.J. | | Deposit date: | 2023-03-24 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.38 Å) | | Cite: | Structure of the MlaC-MlaD complex reveals molecular basis of periplasmic phospholipid transport.
Nat Commun, 15, 2024
|
|
8OJ4
 
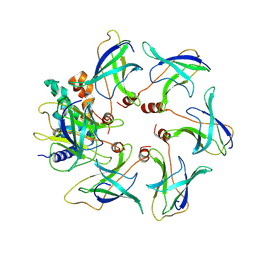 | | Structure of the MlaCD complex (1:6 stoichiometry) | | Descriptor: | Intermembrane phospholipid transport system binding protein MlaC, Intermembrane phospholipid transport system binding protein MlaD | | Authors: | Wotherspoon, P, Bui, S, Sridhar, P, Bergeron, J.R.C, Knowles, T.J. | | Deposit date: | 2023-03-23 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Structure of the MlaC-MlaD complex reveals molecular basis of periplasmic phospholipid transport.
Nat Commun, 15, 2024
|
|
2FNO
 
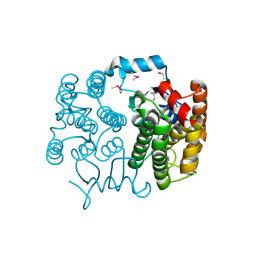 | |
