3WWI
 
 | | Crystal structure of the G136F mutant of the first R-stereoselective -transaminase identified from Arthrobacter sp. KNK168 (FERM-BP-5228) | | Descriptor: | (R)-amine transaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Guan, L.J, Ohtsuka, J, Miyakawa, T, Zhi, Y, Ito, N, Yasohara, Y, Tanokura, M. | | Deposit date: | 2014-06-18 | | Release date: | 2015-08-19 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | A new target region for changing the substrate specificity of amine transaminases.
Sci Rep, 5, 2015
|
|
3WWJ
 
 | | Crystal structure of an engineered sitagliptin-producing transaminase, ATA-117-Rd11 | | Descriptor: | (R)-amine transaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Guan, L.J, Ohtsuka, J, Okai, M, Miyakawa, T, Mase, T, Zhi, Y, Hou, F, Ito, N, Yasohara, Y, Tanokura, M. | | Deposit date: | 2014-06-18 | | Release date: | 2015-08-12 | | Last modified: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A new target region for changing the substrate specificity of amine transaminases.
Sci Rep, 5, 2015
|
|
3X0Y
 
 | | Crystal structure of FMN-bound DszC from Rhodococcus erythropolis D-1 | | Descriptor: | DszC, FLAVIN MONONUCLEOTIDE | | Authors: | Guan, L.J, Lee, W.C, Wang, S.P, Ohtsuka, J, Tanokura, M. | | Deposit date: | 2014-10-23 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of apo-DszC and FMN-bound DszC from Rhodococcus erythropolis D-1.
Febs J., 282, 2015
|
|
3X0X
 
 | | Crystal structure of apo-DszC from Rhodococcus erythropolis D-1 | | Descriptor: | DszC | | Authors: | Guan, L.J, Lee, W.C, Wang, S.P, Ohtsuka, J, Tanokura, M. | | Deposit date: | 2014-10-23 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structures of apo-DszC and FMN-bound DszC from Rhodococcus erythropolis D-1.
Febs J., 282, 2015
|
|
3WWH
 
 | | Crystal structure of the first R-stereoselective -transaminase identified from Arthrobacter sp. KNK168 (FERM-BP-5228) | | Descriptor: | (R)-amine transaminase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Guan, L.J, Ohtsuka, J, Okai, M, Miyakawa, T, Mase, T, Zhi, Y, Ito, N, Yasohara, Y, Tanokura, M. | | Deposit date: | 2014-06-18 | | Release date: | 2015-08-12 | | Last modified: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A new target region for changing the substrate specificity of amine transaminases.
Sci Rep, 5, 2015
|
|
3WIB
 
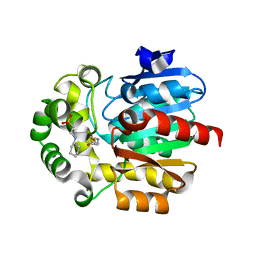 | | Crystal structure of Y109W Mutant Haloalkane Dehalogenase DatA from Agrobacterium tumefaciens C58 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Haloalkane dehalogenase | | Authors: | Guan, L.J, Yabuki, H, Okai, M, Ohtsuka, J, Tanokura, M. | | Deposit date: | 2013-09-09 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the novel haloalkane dehalogenase DatA from Agrobacterium tumefaciens C58 reveals a special halide-stabilizing pair and enantioselectivity mechanism.
Appl.Microbiol.Biotechnol., 98, 2014
|
|
3WI7
 
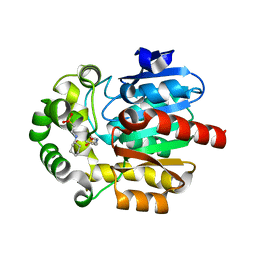 | | Crystal Structure of the Novel Haloalkane Dehalogenase DatA from Agrobacterium tumefaciens C58 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Haloalkane dehalogenase | | Authors: | Guan, L.J, Yabuki, H, Okai, M, Ohtsuka, J, Tanokura, M. | | Deposit date: | 2013-09-06 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the novel haloalkane dehalogenase DatA from Agrobacterium tumefaciens C58 reveals a special halide-stabilizing pair and enantioselectivity mechanism.
Appl.Microbiol.Biotechnol., 98, 2014
|
|
5TLC
 
 | |
