1ZB9
 
 | | Crystal structure of Xylella fastidiosa organic peroxide resistance protein | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, organic hydroperoxide resistance protein | | Authors: | Oliveira, M.A, Guimaraes, B.G, Cussiol, J.R, Medrano, F.J, Vidigal, S.A, Gozzo, F.C, Netto, L.E. | | Deposit date: | 2005-04-07 | | Release date: | 2006-04-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into Enzyme-Substrate Interaction and Characterization of Enzymatic Intermediates of Organic Hydroperoxide Resistance Protein from Xylella fastidiosa.
J.Mol.Biol., 359, 2006
|
|
1ZB8
 
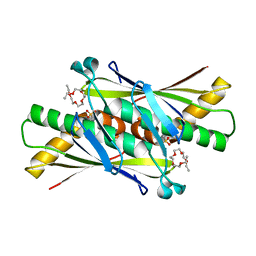 | | Crystal structure of Xylella fastidiosa organic peroxide resistance protein | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, organic hydroperoxide resistance protein | | Authors: | Oliveira, M.A, Guimaraes, B.G, Cussiol, J.R, Medrano, F.J, Vidigal, S.A, Gozzo, F.C, Netto, L.E. | | Deposit date: | 2005-04-07 | | Release date: | 2006-05-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into Enzyme-Substrate Interaction and Characterization of Enzymatic Intermediates of Organic Hydroperoxide Resistance Protein from Xylella fastidiosa.
J.Mol.Biol., 359, 2006
|
|
5D9G
 
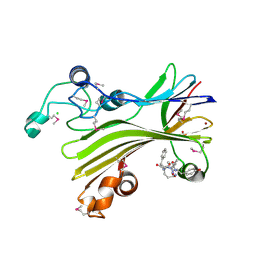 | | Crystal structure of TIPRL, TOR signaling pathway regulator-like, in complex with peptide | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Scorsato, V, Sandy, J, Brandao-Neto, J, Pereira, H.M, Smetana, J.H.C, Aparicio, R. | | Deposit date: | 2015-08-18 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the human Tip41 orthologue, TIPRL, reveals a novel fold and a binding site for the PP2Ac C-terminus.
Sci Rep, 6, 2016
|
|
5VOK
 
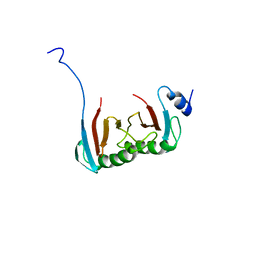 | | Crystal structure of the C7orf59-HBXIP dimer | | Descriptor: | Ragulator complex protein LAMTOR4, Ragulator complex protein LAMTOR5 | | Authors: | Rasheed, N, Nascimento, A.F.Z, Bar-Peled, L, Shen, K, Sabatini, D.M, Aparicio, R, Smetana, J.H.C. | | Deposit date: | 2017-05-03 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | C7orf59/Lamtor4 phosphorylation and structural flexibility modulate Ragulator assembly.
Febs Open Bio, 2019
|
|
6XN2
 
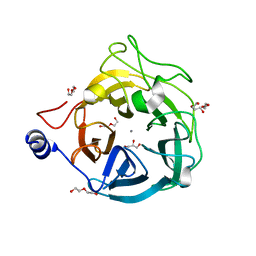 | | Crystal structure of the GH43_1 enzyme from Xanthomonas citri complexed with xylotriose | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Morais, M.A.B, Tonoli, C.C.C, Santos, C.R, Murakami, M.T. | | Deposit date: | 2020-07-02 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Two distinct catalytic pathways for GH43 xylanolytic enzymes unveiled by X-ray and QM/MM simulations.
Nat Commun, 12, 2021
|
|
6XN1
 
 | | Crystal structure of the GH43_1 enzyme from Xanthomonas citri complexed with xylose | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Morais, M.A.B, Tonoli, C.C.C, Santos, C.R, Murakami, M.T. | | Deposit date: | 2020-07-02 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two distinct catalytic pathways for GH43 xylanolytic enzymes unveiled by X-ray and QM/MM simulations.
Nat Commun, 12, 2021
|
|
6XN0
 
 | | Crystal structure of GH43_1 enzyme from Xanthomonas citri | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Morais, M.A.B, Tonoli, C.C.C, Santos, C.R, Murakami, M.T. | | Deposit date: | 2020-07-02 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Two distinct catalytic pathways for GH43 xylanolytic enzymes unveiled by X-ray and QM/MM simulations.
Nat Commun, 12, 2021
|
|
6MOY
 
 | |
6MP7
 
 | | Crystal structure of the E257A mutant of BlMan5B in complex with GlcNAc (soaking) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BlMan5B, CITRATE ANION, ... | | Authors: | Lorizolla-Cordeiro, R, Giuseppe, P.O, Murakami, M.T. | | Deposit date: | 2018-10-05 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | N-glycan Utilization by Bifidobacterium Gut Symbionts Involves a Specialist beta-Mannosidase.
J. Mol. Biol., 431, 2019
|
|
6MP2
 
 | |
6MPA
 
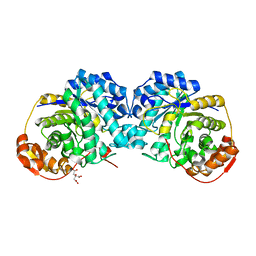 | |
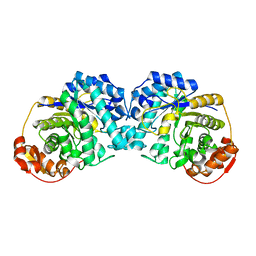
6MPC
 
 | |
6BYI
 
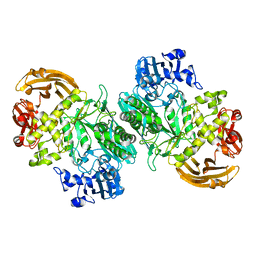 | | Crystal structure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri | | Descriptor: | Beta-mannosidase, beta-D-mannopyranose | | Authors: | Domingues, M.N, Vieira, P.S, Morais, M.A.B, Murakami, M.T. | | Deposit date: | 2017-12-20 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of exo-beta-mannanase activity in the GH2 family.
J. Biol. Chem., 293, 2018
|
|
6D25
 
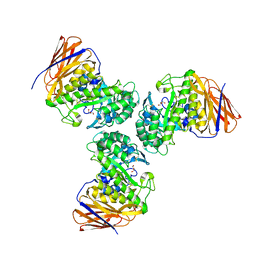 | | Crystal structure of the GH51 arabinofuranosidase from Xanthomonas axonopodis pv. citri | | Descriptor: | Alpha-L-arabinosidase, GLYCEROL | | Authors: | Santos, C.R, Morais, M.A.B, Tonoli, C.C.C, Giuseppe, P.O, Murakami, M.T. | | Deposit date: | 2018-04-13 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The mechanism by which a distinguishing arabinofuranosidase can cope with internal di-substitutions in arabinoxylans.
Biotechnol Biofuels, 11, 2018
|
|
6BYC
 
 | | Crystal structure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri | | Descriptor: | ACETATE ION, Beta-mannosidase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Domingues, M.N, Vieira, P.S, Morais, M.A.B, Murakami, M.T. | | Deposit date: | 2017-12-20 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Structural basis of exo-beta-mannanase activity in the GH2 family.
J. Biol. Chem., 293, 2018
|
|
6BYE
 
 | | Crystal structure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose | | Descriptor: | ACETATE ION, Beta-mannosidase, beta-D-mannopyranose | | Authors: | Domingues, M.N, Vieira, P.S, Morais, M.A.B, Murakami, M.T. | | Deposit date: | 2017-12-20 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.126 Å) | | Cite: | Structural basis of exo-beta-mannanase activity in the GH2 family.
J. Biol. Chem., 293, 2018
|
|
6BYG
 
 | | Crystal structure of the nucleophile mutant (E575A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri | | Descriptor: | Beta-mannosidase, beta-D-mannopyranose | | Authors: | Domingues, M.N, Vieira, P.S, Morais, M.A.B, Murakami, M.T. | | Deposit date: | 2017-12-20 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural basis of exo-beta-mannanase activity in the GH2 family.
J. Biol. Chem., 293, 2018
|
|
7KN8
 
 | | Crystal structure of the GH74 xyloglucanase from Xanthomonas campestris (Xcc1752) | | Descriptor: | 1,2-ETHANEDIOL, Cellulase, IODIDE ION, ... | | Authors: | Araujo, E.A, Vieira, P.S, Murakami, M.T, Polikarpov, I. | | Deposit date: | 2020-11-04 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Xyloglucan processing machinery in Xanthomonas pathogens and its role in the transcriptional activation of virulence factors
Nature Communications, 12, 2021
|
|
7KMN
 
 | |
7KMP
 
 | |
7KMQ
 
 | |
7KMM
 
 | |
7KMO
 
 | |
7KNC
 
 | |
6UB3
 
 | | Crystal structure of a GH128 (subgroup IV) endo-beta-1,3-glucanase from Lentinula edodes (LeGH128_IV) with laminaribiose at the surface-binding site | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, SULFATE ION, ... | | Authors: | Santos, C.R, Lima, E.A, Mandelli, F, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
