6QI5
 
 | | Near Atomic Structure of an Atadenovirus Shows a possible gene duplication event and Intergenera Variations in Cementing Proteins | | Descriptor: | Hexon protein, PIIIa, Penton protein, ... | | Authors: | Condezo, G.N, Marabini, R, Gomez-Blanco, J, SanMartin, C. | | Deposit date: | 2019-01-17 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Near-atomic structure of an atadenovirus reveals a conserved capsid-binding motif and intergenera variations in cementing proteins.
Sci Adv, 7, 2021
|
|
8QTZ
 
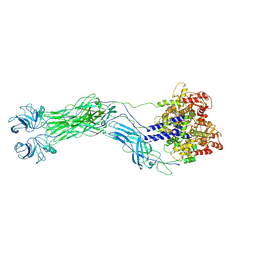 | | Cryo-EM reconstruction of VP5*/VP8* assembly from SA11 Rotavirus Tripsinized Triple Layered Particle | | Descriptor: | Outer capsid protein VP4 | | Authors: | Asensio-Cob, D, Perez-Mata, C, Gomez-Blanco, J, Vargas, J, Rodriguez, J.M, Luque, D. | | Deposit date: | 2023-10-13 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural basis of rotavirus spike proteolytic activation
To Be Published
|
|
3ZUE
 
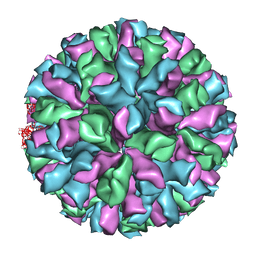 | | Rabbit Hemorrhagic Disease Virus (RHDV)capsid protein | | Descriptor: | CAPSID STRUCTURAL PROTEIN VP60 | | Authors: | Luque, D, Gonzalez, J.M, Gomez-Blanco, J, Marabini, R, Chichon, J, Mena, I, Angulo, I, Carrascosa, J.L, Verdaguer, N, Trus, B.L, Barcena, J, Caston, J.R. | | Deposit date: | 2011-07-18 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.3 Å) | | Cite: | Epitope Insertion at the N-Terminal Molecular Switch of the Rabbit Hemorrhagic Disease Virus T=3 Capsid Protein Leads to Larger T=4 Capsids.
J.Virol., 86, 2012
|
|
3J3I
 
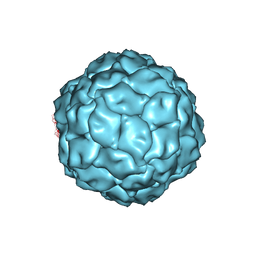 | | Penicillium chrysogenum virus (PcV) capsid structure | | Descriptor: | Capsid protein | | Authors: | Luque, D, Gomez-Blanco, J, Garriga, D, Brilot, A, Gonzalez, J.M, Havens, W.H, Carrascosa, J.L, Trus, B.L, Verdaguer, N, Grigorieff, N, Ghabrial, S.A, Caston, J.R. | | Deposit date: | 2013-03-08 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM near-atomic structure of a dsRNA fungal virus shows ancient structural motifs preserved in the dsRNA viral lineage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8OLE
 
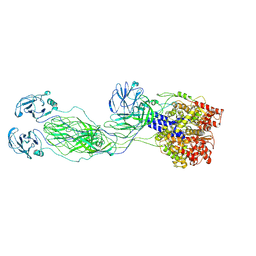 | | Cryo-EM reconstruction of VP4 assembly from SA11 Rotavirus Non-Tripsinized Triple Layered Particle | | Descriptor: | Outer capsid protein VP4 | | Authors: | Asensio-Cob, D, Perez-Mata, C, Gomez-Blanco, J, Vargas, J, Rodriguez, J.M, Luque, D. | | Deposit date: | 2023-03-30 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis of rotavirus spike proteolytic activation
To Be Published
|
|
8OLB
 
 | | SA11 Rotavirus Non-tripsinized Triple Layered Particle | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Inner capsid protein VP2, ... | | Authors: | Asensio-Cob, D, Perez-Mata, C, Gomez-Blanco, J, Vargas, J, Rodriguez, J.M, Luque, D. | | Deposit date: | 2023-03-30 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of rotavirus spike proteolytic activation
To Be Published
|
|
8OLC
 
 | | SA11 Rotavirus Trypsinized Triple Layered Particle | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Inner capsid protein VP2, ... | | Authors: | Asensio-Cob, D, Perez-Mata, C, Gomez-Blanco, J, Vargas, J, Rodriguez, J.M, Luque, D. | | Deposit date: | 2023-03-30 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural basis of rotavirus spike proteolytic activation
To Be Published
|
|
6W7N
 
 | | 30S-Inactive-low-Mg2+ Class A | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S12, ... | | Authors: | Jahagirdar, D, Jha, V, Basu, B, Gomez-Blanco, J, Vargas, J, Ortega, J. | | Deposit date: | 2020-03-19 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy.
Rna, 26, 2020
|
|
6W6K
 
 | | 30S-Activated-high-Mg2+ | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Jahagirdar, D, Jha, V, Basu, B, Gomez-Blanco, J, Vargas, J, Ortega, J. | | Deposit date: | 2020-03-17 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy.
Rna, 26, 2020
|
|
6W77
 
 | | 30S-Inactivated-high-Mg2+ Class A | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Jahagirdar, D, Jha, V, Basu, B, Gomez-Blanco, J, Vargas, J, Ortega, J. | | Deposit date: | 2020-03-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy.
Rna, 26, 2020
|
|
6W7M
 
 | | 30S-Inactive-high-Mg2+ + carbon layer | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Jahagirdar, D, Jha, V, Basu, B, Gomez-Blanco, J, Vargas, J, Ortega, J. | | Deposit date: | 2020-03-19 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy.
Rna, 26, 2020
|
|
6W7W
 
 | | 30S-Inactive-low-Mg2+ Class B | | Descriptor: | 16S rRNA, 30S ribosomal protein S12, 30S ribosomal protein S15, ... | | Authors: | Jahagirdar, D, Jha, V, Basu, B, Gomez-Blanco, J, Vargas, J, Ortega, J. | | Deposit date: | 2020-03-19 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy.
Rna, 26, 2020
|
|
4ZIU
 
 | | Crystal structure of native alpha-2-macroglobulin from Escherichia coli spanning the residues from domain MG7 to the C-terminus. | | Descriptor: | GLYCEROL, NICKEL (II) ION, Uncharacterized lipoprotein YfhM | | Authors: | Garcia-Ferrer, I, Arede, P, Gomez-Blanco, J, Luque, D, Duquerroy, S, Caston, J.R, Goulas, T, Gomis-Ruth, X.F. | | Deposit date: | 2015-04-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional insights into Escherichia coli alpha 2-macroglobulin endopeptidase snap-trap inhibition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZJH
 
 | | Crystal structure of native alpha-2-macroglobulin from Escherichia coli spanning domains NIE-MG1. | | Descriptor: | ACETATE ION, GLYCEROL, alpha-2-Macroglobulin | | Authors: | Garcia-Ferrer, I, Arede, P, Gomez-Blanco, J, Luque, D, Duquerroy, S, Caston, J.R, Goulas, T, Gomis-Ruth, X.F. | | Deposit date: | 2015-04-29 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional insights into Escherichia coli alpha 2-macroglobulin endopeptidase snap-trap inhibition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZJG
 
 | | Crystal structure of native alpha-2-macroglobulin from Escherichia coli spanning domains MG0-NIE-MG1. | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Garcia-Ferrer, I, Arede, P, Gomez-Blanco, J, Luque, D, Duquerroy, S, Caston, J.R, Goulas, T, Gomis-Ruth, X.F. | | Deposit date: | 2015-04-29 | | Release date: | 2015-06-10 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional insights into Escherichia coli alpha 2-macroglobulin endopeptidase snap-trap inhibition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZIQ
 
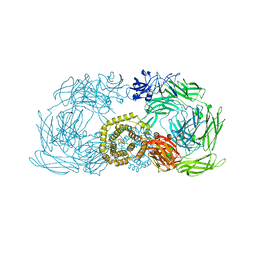 | | Crystal structure of trypsin activated alpha-2-macroglobulin from Escherichia coli. | | Descriptor: | CHLORIDE ION, GLYCEROL, Uncharacterized lipoprotein YfhM | | Authors: | Garcia-Ferrer, I, Arede, P, Gomez-Blanco, J, Luque, D, Duquerroy, S, Caston, J.R, Goulas, T, Gomis-Ruth, X.F. | | Deposit date: | 2015-04-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and functional insights into Escherichia coli alpha 2-macroglobulin endopeptidase snap-trap inhibition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A42
 
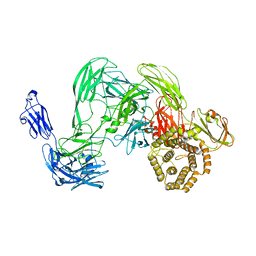 | | Cryo-EM single particle 3D reconstruction of the native conformation of E. coli alpha-2-macroglobulin (ECAM) | | Descriptor: | UNCHARACTERIZED LIPOPROTEIN YFHM | | Authors: | Garcia-Ferrer, I, Arede, P, Gomez-Blanco, J, Luque, D, Duquerroy, S, Caston, J.R, Goulas, T, Gomis-Ruth, F.X. | | Deposit date: | 2015-06-04 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structural and Functional Insights Into Escherichia Coli Alpha2- Macroglobulin Endopeptidase Snap-Trap Inhibition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6NQB
 
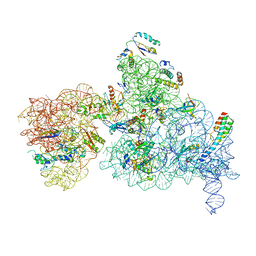 | |
5ND1
 
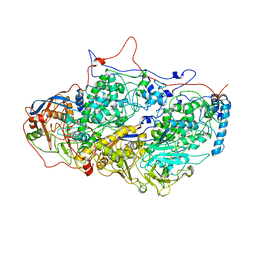 | | Viral evolution results in multiple, surface-allocated enzymatic activities in a fungal double-stranded RNA virus | | Descriptor: | Capsid protein | | Authors: | Mata, C.P, Luque, D, Gomez Blanco, J, Rodriguez, J.M, Suzuki, N, Ghabrial, S.A, Carrascosa, J.L, Trus, B.L, Caston, J.R. | | Deposit date: | 2017-03-07 | | Release date: | 2017-11-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Acquisition of functions on the outer capsid surface during evolution of double-stranded RNA fungal viruses.
PLoS Pathog., 13, 2017
|
|
7MCS
 
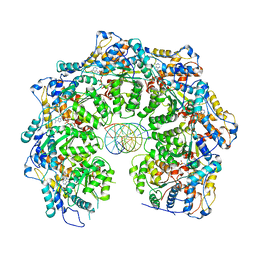 | | Cryo-electron microscopy structure of TnsC(1-503)A225V bound to DNA | | Descriptor: | DNA (5'-D(P*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Shen, Y, Ortega, J, Guarne, A. | | Deposit date: | 2021-04-02 | | Release date: | 2022-02-23 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis for DNA targeting by the Tn7 transposon.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7MBW
 
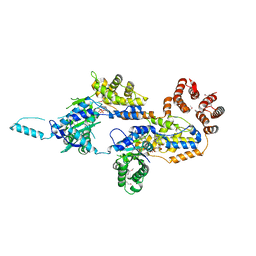 | | Crystal structure of TnsC(1-503)A225V | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transposon Tn7 transposition protein TnsC | | Authors: | Shen, Y, Guarne, A. | | Deposit date: | 2021-04-01 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for DNA targeting by the Tn7 transposon.
Nat.Struct.Mol.Biol., 29, 2022
|
|
