7NXK
 
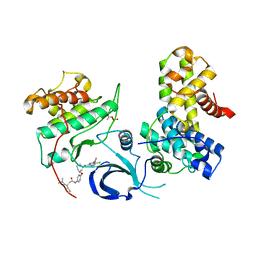 | | Crystal structure of human Cdk12/Cyclin K in complex with the inhibitor BSJ-01-175 | | Descriptor: | (E)-N-[4-[(1R,3R)-3-[[5-chloranyl-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]cyclohexyl]oxyphenyl]-4-(dimethylamino)but-2-enamide, Cyclin-K, Cyclin-dependent kinase 12 | | Authors: | Anand, K, Dust, S, Kaltheuner, I.H, Geyer, M. | | Deposit date: | 2021-03-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-activity relationship study of THZ531 derivatives enables the discovery of BSJ-01-175 as a dual CDK12/13 covalent inhibitor with efficacy in Ewing sarcoma.
Eur.J.Med.Chem., 221, 2021
|
|
2M2F
 
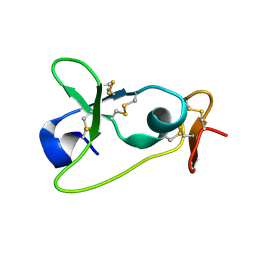 | | The membran-proximal domain of ADAM17 | | Descriptor: | Disintegrin and metalloproteinase domain-containing protein 17 | | Authors: | Duesterhoeft, S, Jung, S, Hung, C, Tholey, A, Soennichsen, F.D, Groetzinger, J, Lorenzen, I. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Membrane-proximal domain of a disintegrin and metalloprotease-17 represents the putative molecular switch of its shedding activity operated by protein-disulfide isomerase.
J.Am.Chem.Soc., 135, 2013
|
|
7NXJ
 
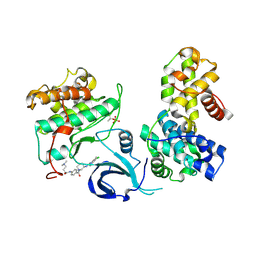 | | Crystal structure of human Cdk13/Cyclin K in complex with the inhibitor THZ531 | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 13, N-[4-[(3R)-3-[[5-chloranyl-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]piperidin-1-yl]carbonylphenyl]-4-(dimethylamino)butanamide | | Authors: | Anand, K, Greifenberg, A.K, Kaltheuner, I.H, Geyer, M. | | Deposit date: | 2021-03-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure-activity relationship study of THZ531 derivatives enables the discovery of BSJ-01-175 as a dual CDK12/13 covalent inhibitor with efficacy in Ewing sarcoma.
Eur.J.Med.Chem., 221, 2021
|
|
2LVL
 
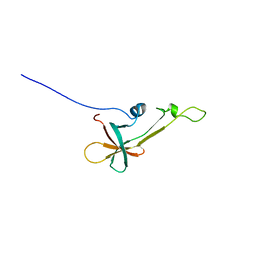 | | NMR Structure the lantibiotic immunity protein SpaI | | Descriptor: | SpaI | | Authors: | Christ, N, Bochmann, S, Gottstein, D, Duchardt-Ferner, E, Hellmich, U.A, Duesterhus, S, Koetter, P, Guentert, P, Entian, K, Woehnert, J. | | Deposit date: | 2012-07-06 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The First Structure of a Lantibiotic Immunity Protein, SpaI from Bacillus subtilis, Reveals a Novel Fold.
J.Biol.Chem., 287, 2012
|
|
2N32
 
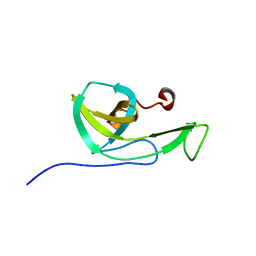 | | NMR solution structure of the N-terminal domain of NisI, a lipoprotein from Lactococcus lactis which confers immunity against nisin | | Descriptor: | Nisin immunity protein | | Authors: | Hacker, C, Christ, N.A, Korn, S, Duchardt-Ferner, E, Hellmich, U.A, Duesterhus, S, Koetter, P, Entian, K, Woehnert, J. | | Deposit date: | 2015-05-21 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of the Lantibiotic Immunity Protein NisI and Its Interactions with Nisin.
J.Biol.Chem., 290, 2015
|
|
2N2E
 
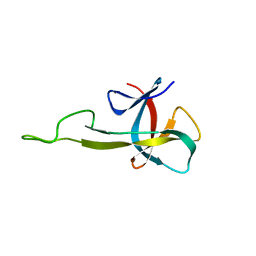 | | NMR solution structure of the C-terminal domain of NisI, a lipoprotein from Lactococcus lactis which confers immunity against nisin | | Descriptor: | Nisin immunity protein | | Authors: | Hacker, C, Christ, N.A, Korn, S, Duchardt-Ferner, E, Hellmich, U.A, Duesterhus, S, Koetter, P, Entian, K, Woehnert, J. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of the Lantibiotic Immunity Protein NisI and Its Interactions with Nisin.
J.Biol.Chem., 290, 2015
|
|
