5M21
 
 | | Crystal structure of hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 with 4-hydroxybenzoate bound | | Descriptor: | FE (III) ION, Hydroquinone dioxygenase large subunit, Hydroquinone dioxygenase small subunit, ... | | Authors: | Ferraroni, M, Da Vela, S, Scozzafava, A, Kolvenbach, B, Corvini, P.F.X. | | Deposit date: | 2016-10-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structures of native hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 and of substrate and inhibitor complexes.
Biochim. Biophys. Acta, 1865, 2017
|
|
5M26
 
 | | Crystal structure of hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 in complex with methylhydroquinone | | Descriptor: | 2-methylbenzene-1,4-diol, FE (III) ION, Hydroquinone dioxygenase large subunit, ... | | Authors: | Ferraroni, M, Da Vela, S, Scozzafava, A, Kolvenbach, B, Corvini, P.F.X. | | Deposit date: | 2016-10-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of native hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 and of substrate and inhibitor complexes.
Biochim. Biophys. Acta, 1865, 2017
|
|
5M22
 
 | | Crystal structure of hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 | | Descriptor: | FE (III) ION, Hydroquinone dioxygenase large subunit, Hydroquinone dioxygenase small subunit | | Authors: | Ferraroni, M, Da Vela, S, Scozzafava, A, Kolvenbach, B, Corvini, P.F.X. | | Deposit date: | 2016-10-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structures of native hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 and of substrate and inhibitor complexes.
Biochim. Biophys. Acta, 1865, 2017
|
|
5M4O
 
 | | Crystal structure of hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 in complex with 4-nitrophenol | | Descriptor: | FE (III) ION, Hydroquinone dioxygenase large subunit, Hydroquinone dioxygenase small subunit, ... | | Authors: | Ferraroni, M, Da Vela, S, Scozzafava, A, Kolvenbach, B, Corvini, P.F.X. | | Deposit date: | 2016-10-18 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structures of native hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 and of substrate and inhibitor complexes.
Biochim. Biophys. Acta, 1865, 2017
|
|
7A9C
 
 | |
8OMR
 
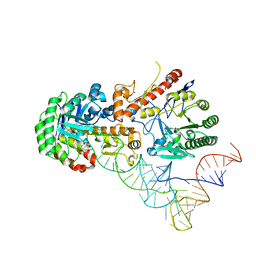 | | Human tRNA guanine transglycosylase (TGT) bound to tRNAAsp | | Descriptor: | 9-DEAZAGUANINE, Queuine tRNA-ribosyltransferase accessory subunit 2, Queuine tRNA-ribosyltransferase catalytic subunit 1, ... | | Authors: | Sievers, K, Neumann, P, Susac, L, Trowitzsch, S, Tampe, R, Ficner, R. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and functional insights into tRNA recognition by human tRNA guanine transglycosylase.
Structure, 32, 2024
|
|
7QDA
 
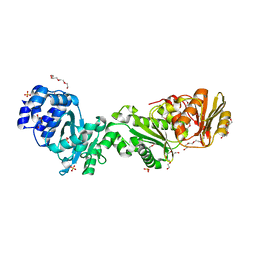 | | Crystal structure of CalpL | | Descriptor: | CalpL, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Schneberger, N, Hagelueken, G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature, 614, 2023
|
|
8B0R
 
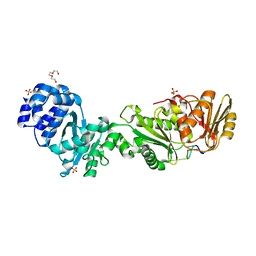 | | Structure of the CalpL/cA4 complex | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), SMODS-associated and fused to various effectors domain-containing protein, SULFATE ION, ... | | Authors: | Schneberger, N, Hagelueken, G. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature, 614, 2023
|
|
8B0U
 
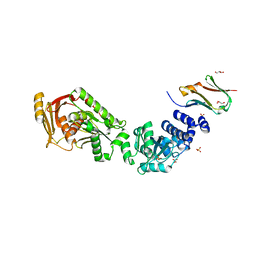 | | Structure of the CalpL/T10 complex | | Descriptor: | CalpT10, GLYCEROL, SAVED domain-containing protein, ... | | Authors: | Schneberger, N, Hagelueken, G. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature, 614, 2023
|
|
