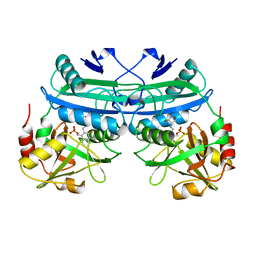
2DAA
 
 | |
2PAN
 
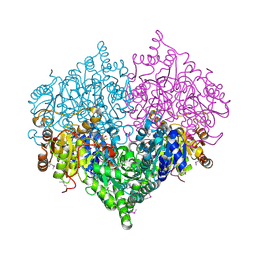 | | Crystal structure of E. coli glyoxylate carboligase | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, FLAVIN-ADENINE DINUCLEOTIDE, Glyoxylate carboligase, ... | | Authors: | Kaplun, A, Chipman, D.M, Barak, Z, Vyazmensky, M, Shaanan, B. | | Deposit date: | 2007-03-27 | | Release date: | 2008-01-01 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Glyoxylate carboligase lacks the canonical active site glutamate of thiamine-dependent enzymes.
Nat.Chem.Biol., 4, 2008
|
|
2F1F
 
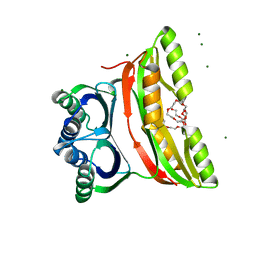 | | Crystal structure of the regulatory subunit of acetohydroxyacid synthase isozyme III from E. coli | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Acetolactate synthase isozyme III small subunit, MAGNESIUM ION, ... | | Authors: | Kaplun, A, Vyazmensky, M, Barak, Z, Chipman, D.M, Shaanan, B. | | Deposit date: | 2005-11-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Regulatory Subunit of Acetohydroxyacid Synthase Isozyme III from Escherichia coli.
J.Mol.Biol., 357, 2006
|
|
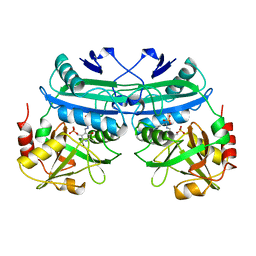
4DAA
 
 | |
3DAA
 
 | |
8BEO
 
 | | Crystal structure of E. coli glyoxylate carboligase mutant I393A with MAP | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2,3-DIMETHOXY-5-METHYL-1,4-BENZOQUINONE, ... | | Authors: | Shaanan, B, Binshtein, E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of E. coli glyoxylate carboligase mutant I393A with MAP
To Be Published
|
|
