8PKM
 
 | | Befiradol-bound serotonin 5-HT1A receptor - Gi Protein Complex | | Descriptor: | (2R)-1-(heptadecanoyloxy)-3-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, (3-chloranyl-4-fluoranyl-phenyl)-[4-fluoranyl-4-[[(5-methylpyridin-2-yl)methylamino]methyl]piperidin-1-yl]methanone, 5-hydroxytryptamine receptor 1A, ... | | Authors: | Schneider, J, Gmeiner, P, Hove, T.T, Rasmussen, T, Boettcher, B. | | Deposit date: | 2023-06-27 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Discovery of a functionally selective serotonin 1A receptor agonist
for the treatment of pain
To Be Published
|
|
8PJK
 
 | | ST171-bound serotonin 5-HT1A receptor - Gi Protein Complex | | Descriptor: | (2R)-1-(heptadecanoyloxy)-3-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 5-hydroxytryptamine receptor 1A, 6-[3-[2-(2-methoxyphenoxy)ethylamino]propoxy]-4~{H}-1,4-benzoxazin-3-one, ... | | Authors: | Schneider, J, Gmeiner, P, Rasmussen, T, Boettcher, B. | | Deposit date: | 2023-06-23 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Discovery of a functionally selective serotonin 1A receptor agonist
for the treatment of pain
To Be Published
|
|
2PWR
 
 | | HIV-1 protease in complex with a carbamoyl decorated pyrrolidine-based inhibitor | | Descriptor: | 4,4'-{(3S,4S)-PYRROLIDINE-3,4-DIYLBIS[(BENZYLIMINO)SULFONYL]}DIBENZAMIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Boettcher, B, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2007-05-12 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Design of C2-Symmetric HIV-1 Protease Inhibitors Based on a Pyrrolidine Scaffold.
J.Med.Chem., 51, 2008
|
|
4ADT
 
 | | Crystal structure of plasmodial PLP synthase | | Descriptor: | PHOSPHATE ION, PYRIDOXINE BIOSYNTHETIC ENZYME PDX1 HOMOLOGUE, PUTATIVE | | Authors: | Guedez, G, Sinning, I, Tews, I. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Assembly of the Eukaryotic Plp-Synthase Complex from Plasmodium and Activation of the Pdx1 Enzyme.
Structure, 20, 2012
|
|
4ADS
 
 | | Crystal structure of plasmodial PLP synthase complex | | Descriptor: | PDX2 PROTEIN, PHOSPHATE ION, PYRIDOXINE BIOSYNTHETIC ENZYME PDX1 HOMOLOGUE, ... | | Authors: | Guedez, G, Sinning, I, Tews, I. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Assembly of the Eukaryotic Plp-Synthase Complex from Plasmodium and Activation of the Pdx1 Enzyme.
Structure, 20, 2012
|
|
4ADU
 
 | | Crystal structure of plasmodial PLP synthase with bound R5P intermediate | | Descriptor: | 1,2-ETHANEDIOL, PYRIDOXINE BIOSYNTHETIC ENZYME PDX1 HOMOLOGUE, PUTATIVE, ... | | Authors: | Guedez, G, Sinning, I, Tews, I. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Assembly of the Eukaryotic Plp-Synthase Complex from Plasmodium and Activation of the Pdx1 Enzyme.
Structure, 20, 2012
|
|
7PZK
 
 | | HBc-WT in complex with Triton X-100 | | Descriptor: | Capsid protein, FRAGMENT OF TRITON X-100 | | Authors: | Makbul, C, Boettcher, B. | | Deposit date: | 2021-10-12 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Binding of a Pocket Factor to Hepatitis B Virus Capsids Changes the Rotamer Conformation of Phenylalanine 97.
Viruses, 13, 2021
|
|
7PZ9
 
 | | HBc-F97L premature secretion phenotype | | Descriptor: | Capsid protein | | Authors: | Makbul, C, Boettcher, B. | | Deposit date: | 2021-10-11 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Binding of a Pocket Factor to Hepatitis B Virus Capsids Changes the Rotamer Conformation of Phenylalanine 97.
Viruses, 13, 2021
|
|
7PZI
 
 | |
7PZL
 
 | | HBc-F97L premature secretion phenotype | | Descriptor: | Capsid protein, FRAGMENT OF TRITON X-100 | | Authors: | Makbul, C, Boettcher, B. | | Deposit date: | 2021-10-12 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Binding of a Pocket Factor to Hepatitis B Virus Capsids Changes the Rotamer Conformation of Phenylalanine 97.
Viruses, 13, 2021
|
|
7PZN
 
 | |
7PZM
 
 | | HBc-P5T in complex with X-100 | | Descriptor: | Capsid protein, FRAGMENT OF TRITON X-100 | | Authors: | Makbul, C, Boettcher, B. | | Deposit date: | 2021-10-12 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Binding of a Pocket Factor to Hepatitis B Virus Capsids Changes the Rotamer Conformation of Phenylalanine 97.
Viruses, 13, 2021
|
|
3J2I
 
 | | Structure of late pre-60S ribosomal subunits with nuclear export factor Arx1 bound at the peptide exit tunnel | | Descriptor: | Eukaryotic translation initiation factor 6, Proliferation-associated protein 2G4 | | Authors: | Bradatsch, B, Leidig, C, Granneman, S, Gnaedig, M, Tollervey, D, Boettcher, B, Beckmann, R, Hurt, E. | | Deposit date: | 2012-10-04 | | Release date: | 2012-11-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.9 Å) | | Cite: | Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
7A46
 
 | | small conductance mechanosensitive channel YbiO | | Descriptor: | Putative transport protein | | Authors: | Flegler, V.J, Rasmussen, A, Rao, S, Wu, N, Zenobi, R, Sansom, M.S.P, Hedrich, R, Rasmussen, T, Boettcher, B. | | Deposit date: | 2020-08-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The MscS-like channel YnaI has a gating mechanism based on flexible pore helices.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7AMV
 
 | | Atomic structure of the poxvirus transcription pre-initiation complex in the initially melted state | | Descriptor: | ATP-dependent helicase VETFS, DNA-directed RNA polymerase, DNA-directed RNA polymerase 147 kDa polypeptide, ... | | Authors: | Grimm, C, Bartuli, J, Fischer, U. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of the complete poxvirus transcription initiation process.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AOF
 
 | | Atomic structure of the poxvirus transcription late pre-initiation complex | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase 147 kDa polypeptide, DNA-directed RNA polymerase 18 kDa subunit, ... | | Authors: | Grimm, C, Bartuli, J, Fischer, U. | | Deposit date: | 2020-10-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of the complete poxvirus transcription initiation process.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AP8
 
 | | Atomic structure of the poxvirus initially transcribing complex in conformation 2 | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase 147 kDa polypeptide, DNA-directed RNA polymerase 18 kDa subunit, ... | | Authors: | Grimm, C, Bartuli, J, Fischer, U. | | Deposit date: | 2020-10-16 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis of the complete poxvirus transcription initiation process.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AOZ
 
 | | Atomic structure of the poxvirus transcription initiation complex in conformation 1 | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase 147 kDa polypeptide, DNA-directed RNA polymerase 18 kDa subunit, ... | | Authors: | Grimm, C, Bartuli, J, Fischer, U. | | Deposit date: | 2020-10-15 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of the complete poxvirus transcription initiation process.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AOH
 
 | | Atomic structure of the poxvirus late initially transcribing complex | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase 147 kDa polypeptide, DNA-directed RNA polymerase 18 kDa subunit, ... | | Authors: | Grimm, C, Bartuli, J, Fischer, U. | | Deposit date: | 2020-10-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of the complete poxvirus transcription initiation process.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AP9
 
 | | Atomic structure of the poxvirus initially transcribing complex in conformation 3 | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase 147 kDa polypeptide, DNA-directed RNA polymerase 18 kDa subunit, ... | | Authors: | Grimm, C, Bartuli, J, Fischer, U. | | Deposit date: | 2020-10-16 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis of the complete poxvirus transcription initiation process.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6RLD
 
 | | STRUCTURE OF THE MECHANOSENSITIVE CHANNEL MSCS EMBEDDED IN THE MEMBRANE BILAYER | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Small-conductance mechanosensitive channel | | Authors: | Rasmussen, T, Flegler, V.J, Rasmussen, A, Boettcher, B. | | Deposit date: | 2019-05-02 | | Release date: | 2019-07-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the Mechanosensitive Channel MscS Embedded in the Membrane Bilayer.
J.Mol.Biol., 431, 2019
|
|
6RFL
 
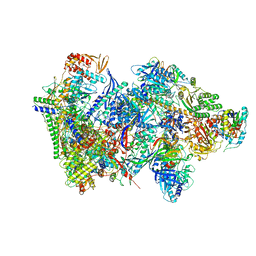 | | Structure of the complete Vaccinia DNA-dependent RNA polymerase complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Grimm, C, Hillen, S.H, Bedenk, K, Bartuli, J, Neyer, S, Zhang, Q, Huettenhofer, A, Erlacher, M, Dienemann, C, Schlosser, A, Urlaub, H, Boettcher, B, Szalay, A.A, Cramer, P, Fischer, U. | | Deposit date: | 2019-04-15 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Vaccinia RNA Polymerase Complexes.
Cell, 179, 2019
|
|
6RX4
 
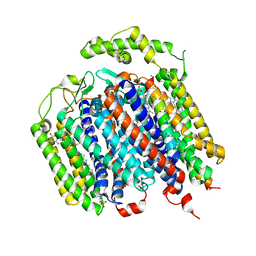 | | THE STRUCTURE OF BD OXIDASE FROM ESCHERICHIA COLI | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome bd-I ubiquinol oxidase subunit 1, ... | | Authors: | Rasmussen, T, Boettcher, B, Thesseling, A, Friedrich, T. | | Deposit date: | 2019-06-07 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Homologous bd oxidases share the same architecture but differ in mechanism.
Nat Commun, 10, 2019
|
|
7OOA
 
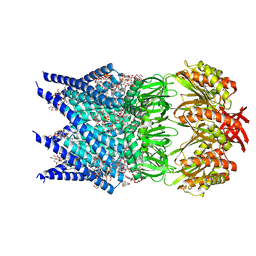 | | Mechanosensitive channel MscS solubilized with LMNG in open conformation with added lipid | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Rasmussen, T, Flegler, V.J, Boettcher, B. | | Deposit date: | 2021-05-27 | | Release date: | 2021-09-01 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanosensitive channel gating by delipidation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OO0
 
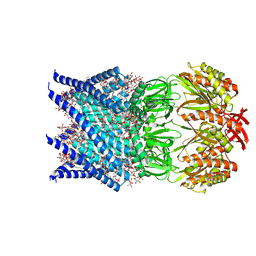 | | Mechanosensitive channel MscS solubilized with DDM in open conformation | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, Mechanosensitive channel MscS | | Authors: | Rasmussen, T, Flegler, V.J, Boettcher, B. | | Deposit date: | 2021-05-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanosensitive channel gating by delipidation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
