1CLB
 
 | |
1IN2
 
 | | Peptide Antagonist of IGFBP1, (i,i+7) Covalently Restrained Analog | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1IN3
 
 | | Peptide Antagonist of IGFBP1, (i,i+8) Covalently Restrained Analog | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1ILP
 
 | |
1GJF
 
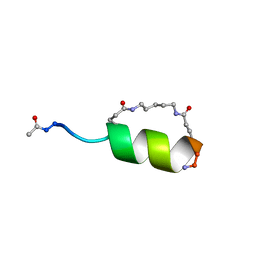 | | Peptide Antagonist of IGFBP1, (i,i+7) Covalently Restrained Analog, Minimized Average Structure | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1GJG
 
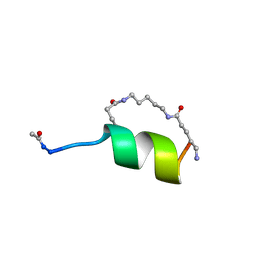 | | Peptide Antagonist of IGFBP1, (i,i+8) Covalently Restrained Analog, Minimized Average Structure | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1RTN
 
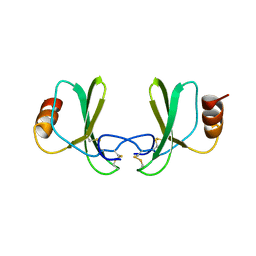 | |
1RTO
 
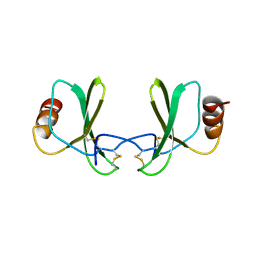 | |
1KVG
 
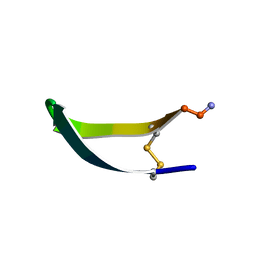 | | EPO-3 beta Hairpin Peptide | | Descriptor: | Protein: EPO-3 Receptor Agonist | | Authors: | Skelton, N.J, Russell, S, de Sauvage, F, Cochran, A.G. | | Deposit date: | 2002-01-25 | | Release date: | 2002-03-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Amino Acid Determinants of beta-Hairpin Conformation in Erythropoeitin
Receptor Agonist Peptides Derived from a Phage Display Library
J.Mol.Biol., 316, 2002
|
|
1KVF
 
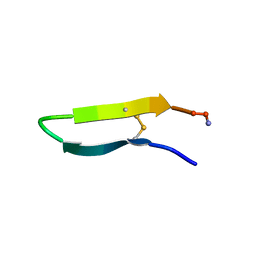 | |
1N7T
 
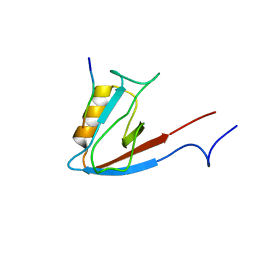 | | ERBIN PDZ domain bound to a phage-derived peptide | | Descriptor: | 99-mer peptide of densin-180-like protein, phage-derived peptide | | Authors: | Skelton, N.J, Koehler, M.F.T, Zobel, K, Wong, W.L, Yeh, S, Pisabarro, M.T, Yin, J.P, Lasky, L.A, Sidhu, S.S. | | Deposit date: | 2002-11-16 | | Release date: | 2003-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Origins of PDZ domain ligand specificity. Structure determination and mutagenesis of the Erbin PDZ domain.
J.Biol.Chem., 278, 2003
|
|
1GNA
 
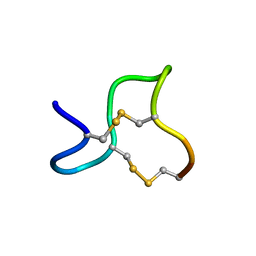 | |
1GNB
 
 | |
1ILQ
 
 | |
1PMX
 
 | | INSULIN-LIKE GROWTH FACTOR-I BOUND TO A PHAGE-DERIVED PEPTIDE | | Descriptor: | IGF-1 ANTAGONIST F1-1, Insulin-like growth factor IB | | Authors: | Skelton, N.J. | | Deposit date: | 2003-06-11 | | Release date: | 2003-10-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Complex with a Phage Display-Derived Peptide Provides Insight into the Function of Insulin-like Growth Factor I
Biochemistry, 42, 2003
|
|
1IMW
 
 | | Peptide Antagonist of IGFBP-1 | | Descriptor: | IGFBP-1 antagonist | | Authors: | Lowman, H.B, Chen, Y.M, Skelton, N.J, Mortensen, D.L, Tomlinson, E.E, Sadick, M.D, Robinson, I.C, Clark, R.G. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1GJE
 
 | | Peptide Antagonist of IGFBP-1, Minimized Average Structure | | Descriptor: | IGFBP-1 antagonist | | Authors: | Lowman, H.B, Chen, Y.M, Skelton, N.J, Mortensen, D.L, Tomlinson, E.E, Sadick, M.D, Robinson, I.C, Clark, R.G. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
2JOA
 
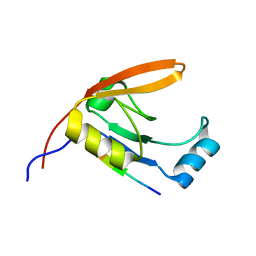 | | HtrA1 bound to an optimized peptide: NMR assignment of PDZ domain and ligand resonances | | Descriptor: | Peptide H1-C1, Serine protease HTRA1 | | Authors: | Runyon, S.T, Zhang, Y, Appleton, B.A, Sazinksy, S.L, Wu, P, Pan, B, Wiesmann, C, Skelton, N.J, Sidhu, S.S. | | Deposit date: | 2007-03-01 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural and functional analysis of the PDZ domains of human HtrA1 and HtrA3
Protein Sci., 16, 2007
|
|
5HIE
 
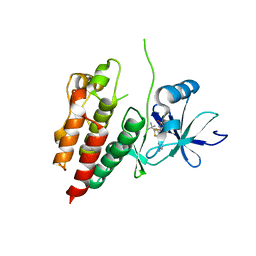 | | BRAF Kinase domain b3aC loop deletion mutant in complex with dabrafenib | | Descriptor: | Dabrafenib, Serine/threonine-protein kinase B-raf | | Authors: | Whalen, D.M, Foster, S.A, Ozen, A, Wongchenko, M, Yin, J, Schaefer, G, Mayfield, J, Chmielecki, J, Stephens, P, Albacker, L, Yan, Y, Song, K, Hatzivassiliou, G, Eigenbrot, C, Yu, C, Shaw, A.S, Manning, G, Skelton, N.J, Hymowitz, S.G, Malek, S. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Activation Mechanism of Oncogenic Deletion Mutations in BRAF, EGFR, and HER2.
Cancer Cell, 29, 2016
|
|
1LE1
 
 | |
1LE0
 
 | |
1LE3
 
 | |
1N09
 
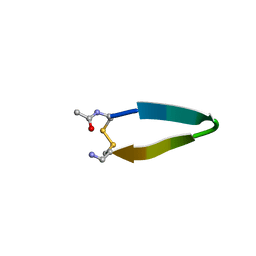 | | A minimal beta-hairpin peptide scaffold for beta-turn display | | Descriptor: | bhpW, disulfide cyclized beta-hairpin peptide | | Authors: | Russell, S.J, Blandl, T, Skelton, N.J, Cochran, A.G. | | Deposit date: | 2002-10-11 | | Release date: | 2003-02-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Stability of cyclic beta-hairpins: asymmetric contributions from side chains of a hydrogen-bonded cross-strand residue pair
J.Am.Chem.Soc., 125, 2003
|
|
1IMX
 
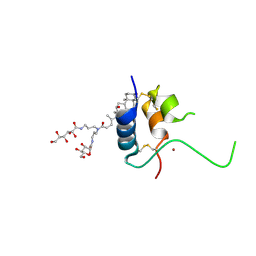 | | 1.8 Angstrom crystal structure of IGF-1 | | Descriptor: | BROMIDE ION, Insulin-like Growth Factor 1A, N,N-BIS(3-D-GLUCONAMIDOPROPYL)DEOXYCHOLAMIDE | | Authors: | Vajdos, F.F, Ultsch, M, Schaffer, M.L, Deshayes, K.D, Liu, J, Skelton, N.J, de Vos, A.M. | | Deposit date: | 2001-05-11 | | Release date: | 2001-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of human insulin-like growth factor-1: detergent binding inhibits binding protein interactions.
Biochemistry, 40, 2001
|
|
3QKM
 
 | | Spirocyclic sulfonamides as AKT inhibitors | | Descriptor: | N-(2-ethoxyethyl)-N-{(2S)-2-hydroxy-3-[(5R)-2-(quinazolin-4-yl)-2,7-diazaspiro[4.5]dec-7-yl]propyl}-2,6-dimethylbenzenesulfonamide, RAC-alpha serine/threonine-protein kinase | | Authors: | Xu, R, Banka, A, Blake, J.F, Mitchell, I.S, Wallace, E.M, Gloor, S.L, Martinson, M, Risom, T, Gross, S.D, Morales, T, Vigers, G.P.A, Brandhuber, B.J, Skelton, N.J. | | Deposit date: | 2011-02-01 | | Release date: | 2011-04-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of spirocyclic sulfonamides as potent Akt inhibitors with exquisite selectivity against PKA.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
