4USN
 
 | | The structure of the immature HIV-1 capsid in intact virus particles at sub-nm resolution | | Descriptor: | P24 | | Authors: | Schur, F.K.M, Hagen, W.J.H, Rumlova, M, Ruml, T, Mueller, B, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2014-07-11 | | Release date: | 2014-11-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structure of the Immature HIV-1 Capsid in Intact Virus Particles at 8.8 A Resolution.
Nature, 517, 2015
|
|
9H1P
 
 | |
8S41
 
 | | The structure of the copia retrotransposon icosahedral capsid (T=9) | | Descriptor: | Copia VLP protein | | Authors: | Klumpe, S, Beck, F, Briggs, J.A.G, Beck, M, Plitzko, J.M. | | Deposit date: | 2024-02-20 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | In-cell structure and snapshots of copia retrotransposons in intact tissue by cryo-ET.
Cell, 188, 2025
|
|
4BZJ
 
 | | The structure of the COPII coat assembled on membranes | | Descriptor: | Protein transport protein SEC13, Protein transport protein SEC31 | | Authors: | Zanetti, G, Prinz, S, Daum, S, Meister, A, Schekman, R, Bacia, K, Briggs, J.A.G. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (40 Å) | | Cite: | The Structure of the Copii Transport-Vesicle Coat Assembled on Membranes
Elife, 2, 2013
|
|
5AHV
 
 | | Cryo-EM structure of helical ANTH and ENTH tubules on PI(4,5)P2-containing membranes | | Descriptor: | ANTH DOMAIN OF ENDOCYTIC ADAPTOR SLA2, ENTH DOMAIN OF EPSIN ENT1 | | Authors: | Skruzny, M, Desfosses, A, Prinz, S, Dodonova, S.O, Gieras, A, Uetrecht, C, Jakobi, A.J, Abella, M, Hagen, W.J.H, Schulz, J, Meijers, R, Rybin, V, Briggs, J.A.G, Sachse, C, Kaksonen, M. | | Deposit date: | 2015-02-10 | | Release date: | 2015-05-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (13.6 Å) | | Cite: | An Organized Co-Assembly of Clathrin Adaptors is Essential for Endocytosis.
Dev.Cell, 33, 2015
|
|
6Z5L
 
 | |
6R23
 
 | | The structure of a Ty3 retrotransposon capsid C-terminal domain dimer | | Descriptor: | Transposon Ty3-I Gag-Pol polyprotein | | Authors: | Dodonova, S.O, Prinz, S, Bilanchone, V, Sandmeyer, S, Briggs, J.A.G. | | Deposit date: | 2019-03-15 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure of the Ty3/Gypsy retrotransposon capsid and the evolution of retroviruses.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6R22
 
 | | The structure of a Ty3 retrotransposon capsid N-terminal domain dimer | | Descriptor: | Transposon Ty3-I Gag-Pol polyprotein | | Authors: | Dodonova, S.O, Prinz, S, Bilanchone, V, Sandmeyer, S, Briggs, J.A.G. | | Deposit date: | 2019-03-15 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structure of the Ty3/Gypsy retrotransposon capsid and the evolution of retroviruses.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6R24
 
 | | The structure of a Ty3 retrotransposon icosahedral capsid | | Descriptor: | Transposon Ty3-I Gag-Pol polyprotein | | Authors: | Dodonova, S.O, Prinz, S, Bilanchone, V, Sandmeyer, S, Briggs, J.A.G. | | Deposit date: | 2019-03-15 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structure of the Ty3/Gypsy retrotransposon capsid and the evolution of retroviruses.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8RB4
 
 | |
8RB5
 
 | |
8RB3
 
 | |
8RB7
 
 | |
9CRI
 
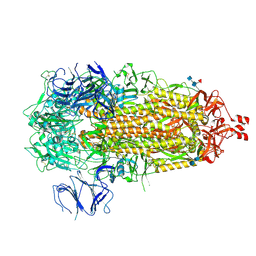 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: Mu (B.1.621) variant 3 closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ke, Z, Briggs, J.A.G. | | Deposit date: | 2024-07-22 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Embo J., 43, 2024
|
|
9CRE
 
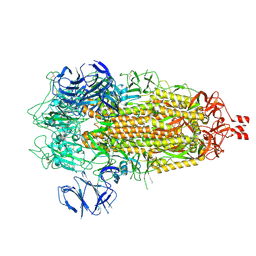 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: Alpha (B.1.1.7) variant 3 closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ke, Z, Croll, T.I, Briggs, J.A.G. | | Deposit date: | 2024-07-22 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Embo J., 43, 2024
|
|
9CRC
 
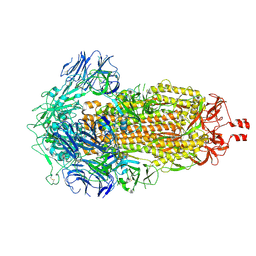 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: B.1 variant 3 closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ke, Z, Croll, T.I, Briggs, J.A.G. | | Deposit date: | 2024-07-22 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Embo J., 43, 2024
|
|
9CRF
 
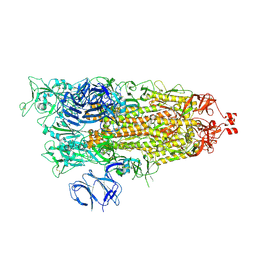 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: Alpha (B.1.1.7) variant 1 open RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ke, Z, Croll, T.I, Briggs, J.A.G. | | Deposit date: | 2024-07-22 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Embo J., 43, 2024
|
|
9CRG
 
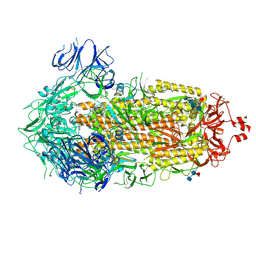 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: Gamma (P.1) variant 3 closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ke, Z, Kotecha, A, Briggs, J.A.G. | | Deposit date: | 2024-07-22 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Embo J., 43, 2024
|
|
9CRD
 
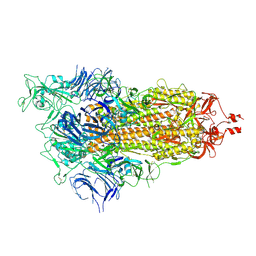 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: B.1 variant 1 open RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ke, Z, Croll, T.I, Briggs, J.A.G. | | Deposit date: | 2024-07-22 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Embo J., 43, 2024
|
|
9CRH
 
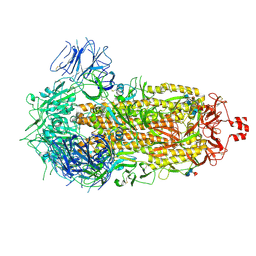 | |
4D1K
 
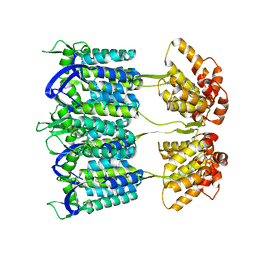 | | Cryo-electron microscopy of tubular arrays of HIV-1 Gag resolves structures essential for immature virus assembly. | | Descriptor: | GAG PROTEIN | | Authors: | Bharat, T.A.M, Castillo-Menendez, L.R, Hagen, W.J.H, Lux, V, Igonet, S, Schorb, M, Schur, F.K.M, Krauesslich, H.G, Briggs, J.A.G. | | Deposit date: | 2014-05-02 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Cryo-Electron Microscopy of Tubular Arrays of HIV-1 Gag Resolves Structures Essential for Immature Virus Assembly.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4COP
 
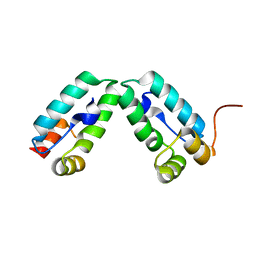 | | HIV-1 capsid C-terminal domain mutant (Y169S) | | Descriptor: | CAPSID PROTEIN P24 | | Authors: | Bharat, T.A.M, Castillo-Menendez, L.R, Hagen, W.J.H, Lux, V, Igonet, S, Schorb, M, Schur, F.K.M, Krausslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2014-01-29 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cryo-Electron Microscopy of Tubular Arrays of HIV-1 Gag Resolves Structures Essential for Immature Virus Assembly.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4COC
 
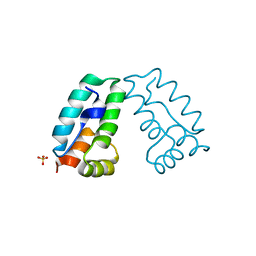 | | HIV-1 capsid C-terminal domain mutant (Y169L) | | Descriptor: | CAPSID PROTEIN P24, SULFATE ION | | Authors: | Bharat, T.A.M, Castillo-Menendez, L.R, Hagen, W.J.H, Lux, V, Igonet, S, Schorb, M, Schur, F.K.M, Krausslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2014-01-28 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Cryo-Electron Microscopy of Tubular Arrays of HIV-1 Gag Resolves Structures Essential for Immature Virus Assembly.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7XU2
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU0
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-211 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
