1P63
 
 | | Human Acidic Fibroblast Growth Factor. 140 Amino Acid Form with Amino Terminal His Tag and Leu111 Replaced with Ile (L111I) | | Descriptor: | ACIDIC FIBROBLAST GROWTH FACTOR, FORMIC ACID, SULFATE ION | | Authors: | Brych, S.R, Kim, J, Logan, T.M, Blaber, M. | | Deposit date: | 2003-04-28 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Accommodation of a highly symmetric core within a symmetric protein
superfold
Protein Sci., 12, 2003
|
|
1MMX
 
 | | Crystal structure of galactose mutarotase from Lactococcus lactis complexed with D-fucose | | Descriptor: | Aldose 1-epimerase, SODIUM ION, alpha-L-fucopyranose | | Authors: | Thoden, J.B, Kim, J, Raushel, F.M, Holden, H.M. | | Deposit date: | 2002-09-04 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis.
J.Biol.Chem., 277, 2002
|
|
3C2G
 
 | | Crystal complex of SYS-1/POP-1 at 2.5A resolution | | Descriptor: | Pop-1 8-residue peptide, Sys-1 protein | | Authors: | Liu, J, Phillips, B.T, Amaya, M.F, Kimble, J, Xu, W. | | Deposit date: | 2008-01-24 | | Release date: | 2008-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The C. elegans SYS-1 protein is a bona fide beta-catenin.
Dev.Cell, 14, 2008
|
|
3C2H
 
 | | Crystal Structure of SYS-1 at 2.6A resolution | | Descriptor: | CITRATE ANION, GLYCEROL, Sys-1 protein | | Authors: | Liu, J, Phillips, B.T, Amaya, M.F, Kimble, J, Xu, W. | | Deposit date: | 2008-01-25 | | Release date: | 2008-05-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The C. elegans SYS-1 protein is a bona fide beta-catenin.
Dev.Cell, 14, 2008
|
|
5XVD
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an air-oxidized condition | | Descriptor: | FE3-S4 CLUSTER, FE4-S4-O CLUSTER, GLYCEROL, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5XVB
 
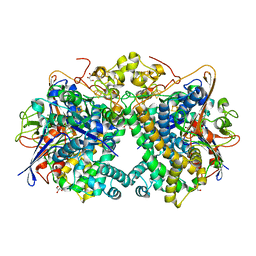 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an H2-reduced condition | | Descriptor: | FE3-S4 CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5XVC
 
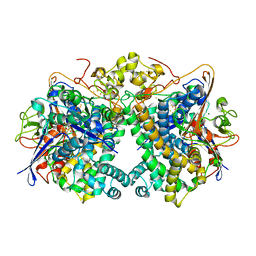 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in a ferricyanide-oxidized condition | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, FE4-S4-O CLUSTER, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5XYK
 
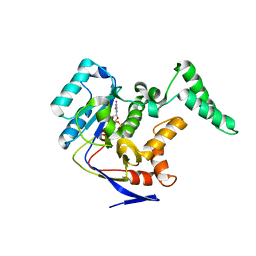 | | Structure of Transferase | | Descriptor: | ARGININE, MANGANESE (II) ION, Putative cytoplasmic protein, ... | | Authors: | Park, J.B, Yoo, Y, Kim, J, Cho, H.S. | | Deposit date: | 2017-07-09 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure of Transferase
To Be Published
|
|
5XZ2
 
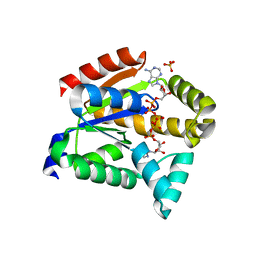 | | Crystal structure of adenylate kinase | | Descriptor: | Adenylate kinase isoenzyme 1, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, SULFATE ION | | Authors: | Moon, S, Bae, E, Kim, J. | | Deposit date: | 2017-07-11 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analyses of adenylate kinases from Antarctic and tropical fishes for understanding cold adaptation of enzymes
Sci Rep, 7, 2017
|
|
5YCB
 
 | |
5YCD
 
 | |
5YCC
 
 | |
5YCF
 
 | |
5X6L
 
 | |
6AI4
 
 | |
5XRU
 
 | |
5W4A
 
 | | C. japonica N-domain | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IMIDAZOLE, ... | | Authors: | Aoki, S.T, Bingman, C.A, Kimble, J. | | Deposit date: | 2017-06-09 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | C. elegans germ granules require both assembly and localized regulators for mRNA repression.
Nat Commun, 12, 2021
|
|
5W4D
 
 | | C. japonica N-domain, Selenomethionine mutant | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, CHLORIDE ION, ... | | Authors: | Aoki, S.T, Bingman, C.A, Kimble, J. | | Deposit date: | 2017-06-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | C. elegans germ granules require both assembly and localized regulators for mRNA repression.
Nat Commun, 12, 2021
|
|
3V74
 
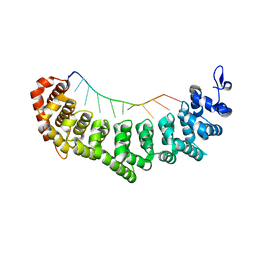 | | crystal structure of FBF-2 in complex with gld-1 FBEa13 RNA | | Descriptor: | Fem-3 mRNA-binding factor 2, RNA (5'-R(*UP*CP*AP*UP*GP*UP*GP*CP*CP*AP*UP*AP*C)-3') | | Authors: | Wang, Y, Qiu, C, Kershner, A, Holley, C.P, Wilinski, D, Keles, S, Kimble, J, Wickens, M, Hall, T.M.T. | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Divergence of PUF protein specificity through variations in an RNA-binding pocket
J.Biol.Chem., 2012
|
|
3V6Y
 
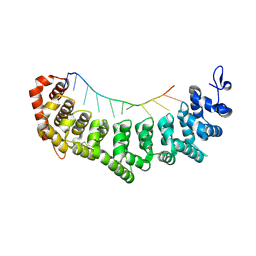 | | crystal structure of FBF-2 in complex with a mutant gld-1 FBEa13 RNA | | Descriptor: | Fem-3 mRNA-binding factor 2, RNA (5'-R(*UP*AP*CP*UP*GP*UP*GP*CP*CP*AP*UP*AP*C)-3') | | Authors: | Qiu, C, Kershner, A, Wang, Y, Holley, C.H, Wilinski, D, Keles, S, Kimble, J, Wickens, M, Hall, T.M.T. | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Divergence of PUF protein specificity through variations in an RNA-binding pocket
J.Biol.Chem., 2012
|
|
3V71
 
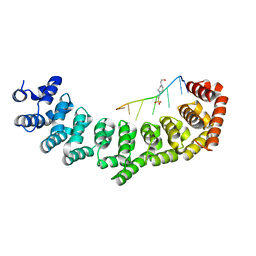 | | Crystal structure of PUF-6 in complex with 5BE13 RNA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Puf (Pumilio/fbf) domain-containing protein 7, confirmed by transcript evidence, ... | | Authors: | Qiu, C, Kershner, A, Wang, Y, Holley, C.H, Wilinski, D, Keles, S, Kimble, J, Wickens, M, Hall, T.M.T. | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Divergence of PUF protein specificity through variations in an RNA-binding pocket
J.Biol.Chem., 2012
|
|
4Y0C
 
 | | The structure of Arabidopsis ClpT2 | | Descriptor: | CHLORIDE ION, Clp protease-related protein At4g12060, chloroplastic, ... | | Authors: | Kimber, M.S, Schultz, L. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Structures, Functions, and Interactions of ClpT1 and ClpT2 in the Clp Protease System of Arabidopsis Chloroplasts.
Plant Cell, 27, 2015
|
|
4Y0B
 
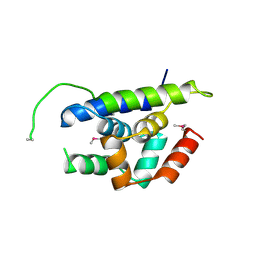 | | The structure of Arabidopsis ClpT1 | | Descriptor: | CHLORIDE ION, Double Clp-N motif protein | | Authors: | Kimber, M.S, Schultz, L. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-13 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures, Functions, and Interactions of ClpT1 and ClpT2 in the Clp Protease System of Arabidopsis Chloroplasts.
Plant Cell, 27, 2015
|
|
6ICI
 
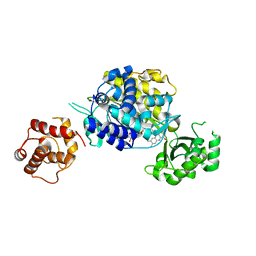 | | Crystal structure of human MICAL3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, [F-actin]-monooxygenase MICAL3 | | Authors: | Hwang, K.Y, Kim, J.S. | | Deposit date: | 2018-09-06 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic insights into flavin-containing monooxygenase and calponin-homology domains in human MICAL3.
Iucrj, 7, 2020
|
|
2NBO
 
 | |
