7KJQ
 
 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to Picloram | | Descriptor: | Picloram, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-26 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KH3
 
 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to Indole 3 propionic acid | | Descriptor: | INDOLYLPROPIONIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-19 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KUA
 
 | | Crystal structure of the MarR family transcriptional regulator from Pseudomonas putida bound to Indole 3 acetic acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Transcriptional regulator, MarR family | | Authors: | Walton, W.G, Lietzan, A.D, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KK0
 
 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to Catechol | | Descriptor: | CATECHOL, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-27 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KKC
 
 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to 5-Hydroxyindoleacetic acid | | Descriptor: | (5-hydroxy-1H-indol-3-yl)acetic acid, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-27 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KRH
 
 | |
7KJL
 
 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to Salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-26 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KKI
 
 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to 2,4-Dichlorophenoxyacetic acid | | Descriptor: | (2,4-DICHLOROPHENOXY)ACETIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-27 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KIG
 
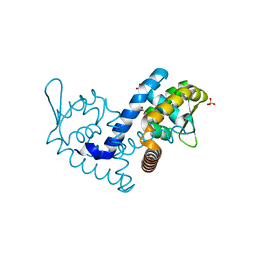 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to Indole-3-butyric acid | | Descriptor: | 3-INDOLEBUTYRIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KYM
 
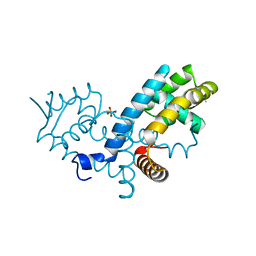 | |
7L19
 
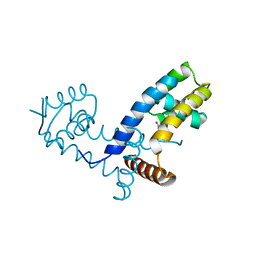 | | Crystal structure of the MarR family transcriptional regulator from Enterobacter soli strain LF7 bound to Indole 3 acetic acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, MarR family transcriptional regulator, NICKEL (II) ION | | Authors: | Lietzan, A.D, Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7L1I
 
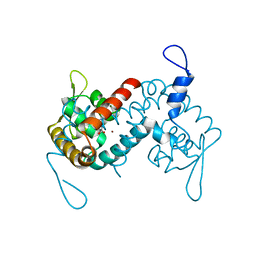 | | Crystal structure of the MarR family transcriptional regulator from Acineotobacter baumannii bound to Indole 3 acetic acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, MarR family multidrug resistance pump transcriptional regulator, NICKEL (II) ION | | Authors: | Walton, W.G, Lietzan, A.D, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-12-14 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
1ATH
 
 | |
3TEC
 
 | | CALCIUM BINDING TO THERMITASE. CRYSTALLOGRAPHIC STUDIES OF THERMITASE AT 0, 5 AND 100 MM CALCIUM | | Descriptor: | CALCIUM ION, EGLIN C, THERMITASE | | Authors: | Gros, P, Kalk, K.H, Hol, W.G.J. | | Deposit date: | 1990-10-26 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Calcium binding to thermitase. Crystallographic studies of thermitase at 0, 5, and 100 mM calcium.
J.Biol.Chem., 266, 1991
|
|
3CVL
 
 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to T. brucei Phosphofructokinase (PFK) PTS1 peptide | | Descriptor: | Peroxisome targeting signal 1 receptor PEX5, T. brucei PFK PTS1 peptide Ac-HEELAKL | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
3CVQ
 
 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to PTS1 peptide (7-SKL) | | Descriptor: | GLYCEROL, PTS1 peptide 7-SKL (Ac-SNRWSKL), Peroxisome targeting signal 1 receptor PEX5 | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
1LT5
 
 | |
3CVP
 
 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to PTS1 peptide (10-SKL) | | Descriptor: | 10-SKL PTS1 peptide Ac-GTLSNRASKL, Peroxisome targeting signal 1 receptor PEX5 | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
3CVN
 
 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to T. brucei Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) PTS1 peptide | | Descriptor: | 1,2-ETHANEDIOL, Peroxisome targeting signal 1 receptor, T. brucei GAPDH PTS1 peptide Ac-DRDAAKL | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
3CFI
 
 | | Nanobody-aided structure determination of the EPSI:EPSJ pseudopilin heterdimer from Vibrio Vulnificus | | Descriptor: | CHLORIDE ION, Nanobody NBEPSIJ_11, Type II secretory pathway, ... | | Authors: | Lam, A.Y, Pardon, E, Korotkov, K.V, Steyaert, J, Hol, W.G.J. | | Deposit date: | 2008-03-03 | | Release date: | 2009-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Nanobody-aided structure determination of the EpsI:EpsJ pseudopilin heterodimer from Vibrio vulnificus.
J.Struct.Biol., 166, 2009
|
|
3CI0
 
 | |
3OSS
 
 | | The crystal structure of enterotoxigenic Escherichia coli GspC-GspD complex from the type II secretion system | | Descriptor: | CALCIUM ION, CHLORIDE ION, TYPE 2 SECRETION SYSTEM, ... | | Authors: | Korotkov, K.V, Pruneda, J, Hol, W.G.J. | | Deposit date: | 2010-09-09 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural and functional studies on the interaction of GspC and GspD in the type II secretion system.
Plos Pathog., 7, 2011
|
|
3CV0
 
 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to T. brucei Phosphoglucoisomerase (PGI) PTS1 peptide | | Descriptor: | 1,2-ETHANEDIOL, Peroxisome targeting signal 1 receptor PEX5, T. brucei PGI PTS1 peptide Ac-FNELSHL | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-17 | | Release date: | 2008-06-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
1LT3
 
 | | HEAT-LABILE ENTEROTOXIN DOUBLE MUTANT N40C/G166C | | Descriptor: | HEAT-LABILE ENTEROTOXIN, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Van Den Akker, F, Hol, W.G.J. | | Deposit date: | 1997-04-12 | | Release date: | 1997-07-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of heat-labile enterotoxin from Escherichia coli with increased thermostability introduced by an engineered disulfide bond in the A subunit.
Protein Sci., 6, 1997
|
|
1LT4
 
 | | HEAT-LABILE ENTEROTOXIN MUTANT S63K | | Descriptor: | HEAT-LABILE ENTEROTOXIN, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Van Den Akker, F, Hol, W.G.J. | | Deposit date: | 1997-04-14 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a non-toxic mutant of heat-labile enterotoxin, which is a potent mucosal adjuvant.
Protein Sci., 6, 1997
|
|
