4DN6
 
 | |
2A1F
 
 | |
2A62
 
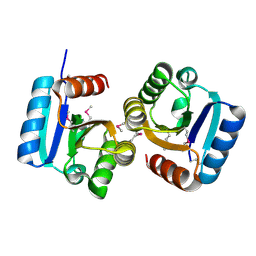
 | | Crystal structure of mouse cadherin-8 EC1-3 | | Descriptor: | CALCIUM ION, Cadherin-8 | | Authors: | Patel, S.D, Ciatto, C, Chen, C.P, Bahna, F, Arkus, N, Schieren, I, Jessell, T.M, Honig, B, Price, S.R, Shapiro, L. | | Deposit date: | 2005-07-01 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Type II cadherin ectodomain structures: implications for classical cadherin specificity.
Cell(Cambridge,Mass.), 124, 2006
|
|
2A4C
 
 | | Crystal structure of mouse cadherin-11 EC1 | | Descriptor: | Cadherin-11 | | Authors: | Patel, S.D, Ciatto, C, Chen, C.P, Bahna, F, Schieren, I, Rajebhosale, M, Jessell, T.M, Honig, B, Price, S.R, Shapiro, L. | | Deposit date: | 2005-06-28 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Type II cadherin ectodomain structures: implications for classical cadherin specificity.
Cell(Cambridge,Mass.), 124, 2006
|
|
3LNE
 
 | |
3LNF
 
 | |
3LNI
 
 | |
3LNH
 
 | |
3LNG
 
 | |
1RKN
 
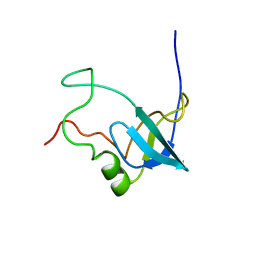 | | Solution structure of 1-110 fragment of Staphylococcal Nuclease with G88W mutation | | Descriptor: | Thermonuclease | | Authors: | Liu, D.S, Feng, Y.G, Ye, K.Q, Shan, L, Wang, J.F. | | Deposit date: | 2003-11-22 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
1U6L
 
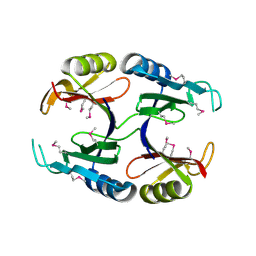 | |
1R3D
 
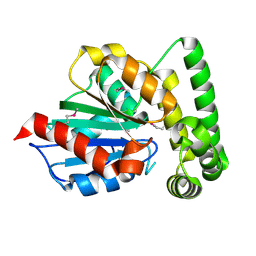 | |
1TR9
 
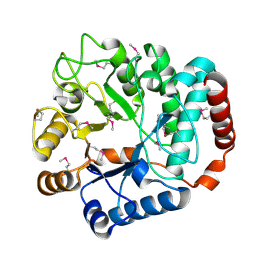 | |
1U6M
 
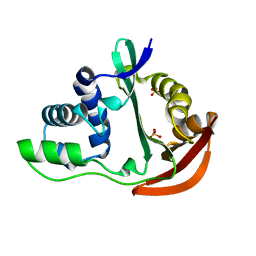 | | The crystal structure of acetyltransferase | | Descriptor: | SULFATE ION, acetyltransferase, GNAT family | | Authors: | Min, T, Gorman, J, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-30 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of acetyltransferase, GNAT family from Enterococcus faecalis
To be Published
|
|
3MW3
 
 | | Crystal structure of beta-neurexin 2 with the splice insert 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neurexin-2-beta | | Authors: | Jin, X, Shapiro, L. | | Deposit date: | 2010-05-05 | | Release date: | 2010-07-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Splice Form Dependence of beta-Neurexin/Neuroligin Binding Interactions.
Neuron, 67, 2010
|
|
3MW4
 
 | | Crystal structure of beta-neurexin 3 without the splice insert 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neurexin-2-beta, ... | | Authors: | Jin, X, Shapiro, L. | | Deposit date: | 2010-05-05 | | Release date: | 2010-07-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Splice Form Dependence of beta-Neurexin/Neuroligin Binding Interactions.
Neuron, 67, 2010
|
|
3MW2
 
 | | Crystal structure of beta-neurexin 1 with the splice insert 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Neurexin-1-alpha, PHOSPHATE ION | | Authors: | Jin, X, Shapiro, L. | | Deposit date: | 2010-05-05 | | Release date: | 2010-07-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Splice Form Dependence of beta-Neurexin/Neuroligin Binding Interactions.
Neuron, 67, 2010
|
|
1S31
 
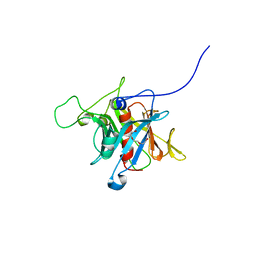 | | Crystal Structure Analysis of the human Tub protein (isoform a) spanning residues 289 through 561 | | Descriptor: | TRIETHYLENE GLYCOL, tubby isoform a | | Authors: | Boutboul, S, Carroll, K.J, Basdevant, A, Gomez, C, Nandrot, E, Clement, K, Shapiro, L, Abitbol, M. | | Deposit date: | 2004-01-12 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | A novel human obesity and sensory deficit syndrome resulting from a mutation in the TUB gene
To be Published
|
|
1TT7
 
 | |
1TVL
 
 | |
1TO3
 
 | |
1T5J
 
 | |
3LND
 
 | |
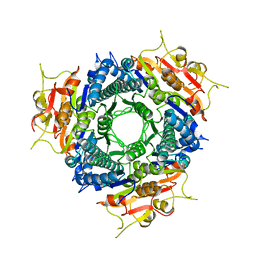
1S80
 
 | | Structure of Serine Acetyltransferase from Haemophilis influenzae Rd | | Descriptor: | Serine acetyltransferase | | Authors: | Gorman, J, Gogos, A, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-01-30 | | Release date: | 2004-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of serine acetyltransferase from Haemophilus influenzae Rd.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1TSJ
 
 | |
