2FDW
 
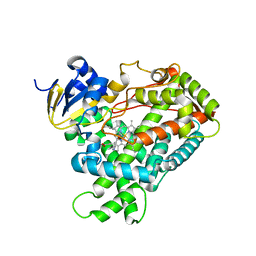 | | Crystal Structure Of Human Microsomal P450 2A6 with the inhibitor (5-(Pyridin-3-yl)furan-2-yl)methanamine bound | | Descriptor: | (5-(PYRIDIN-3-YL)FURAN-2-YL)METHANAMINE, Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yano, J.K, Stout, C.D, Johnson, E.F. | | Deposit date: | 2005-12-14 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthetic Inhibitors of Cytochrome P-450 2A6: Inhibitory Activity, Difference Spectra, Mechanism of Inhibition, and Protein Cocrystallization.
J.Med.Chem., 49, 2006
|
|
2FDU
 
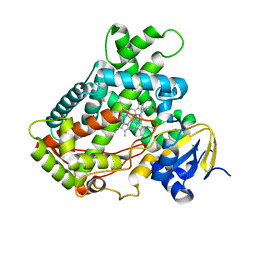 | | Microsomal P450 2A6 with the inhibitor N,N-Dimethyl(5-(pyridin-3-yl)furan-2-yl)methanamine bound | | Descriptor: | Cytochrome P450 2A6, N,N-DIMETHYL(5-(PYRIDIN-3-YL)FURAN-2-YL)METHANAMINE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yano, J.K, Stout, C.D, Johnson, E.F. | | Deposit date: | 2005-12-14 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthetic Inhibitors of Cytochrome P-450 2A6: Inhibitory Activity, Difference Spectra, Mechanism of Inhibition, and Protein Cocrystallization.
J.Med.Chem., 49, 2006
|
|
2L83
 
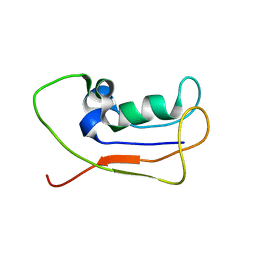 | | A protein from Haloferax volcanii | | Descriptor: | Small archaeal modifier protein 1 | | Authors: | Zhang, W, Liao, S, Fan, K, Tu, X. | | Deposit date: | 2011-01-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ionic strength-dependent conformations of a ubiquitin-like small archaeal modifier protein (SAMP1) from Haloferax volcanii.
Protein Sci., 22, 2013
|
|
2FHS
 
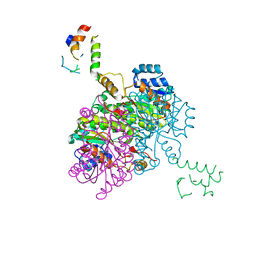 | | Structure of Acyl Carrier Protein Bound to FabI, the Enoyl Reductase from Escherichia Coli | | Descriptor: | Acyl carrier protein, enoyl-[acyl-carrier-protein] reductase, NADH-dependent | | Authors: | Kolappan, S, Novichenok, P, Rafi, S, Simmerling, C, Tonge, P.J, Kisker, C. | | Deposit date: | 2005-12-27 | | Release date: | 2006-10-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Acyl Carrier Protein Bound to FabI, the FASII Enoyl Reductase from Escherichia coli.
J.Biol.Chem., 281, 2006
|
|
2L42
 
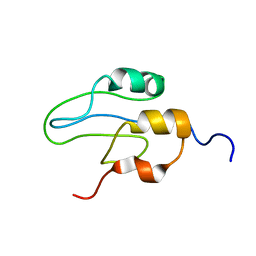 | |
2H2D
 
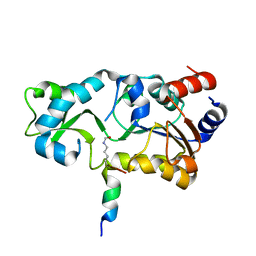 | |
2H2F
 
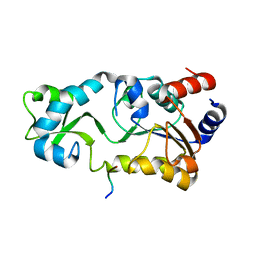 | |
6O3I
 
 | | Crystal Structure of Human IDO1 bound to navoximod (NLG-919) | | Descriptor: | Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE, trans-4-{(1R)-2-[(5S)-6-fluoro-5H-imidazo[5,1-a]isoindol-5-yl]-1-hydroxyethyl}cyclohexan-1-ol | | Authors: | Harris, S.F, Oh, A. | | Deposit date: | 2019-02-26 | | Release date: | 2019-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of Clinical Candidate (1R,4r)-4-((R)-2-((S)-6-Fluoro-5H-imidazo[5,1-a]isoindol-5-yl)-1-hydroxyethyl)cyclohexan-1-ol (Navoximod), a Potent and Selective Inhibitor of Indoleamine 2,3-Dioxygenase 1.
J.Med.Chem., 62, 2019
|
|
6KIG
 
 | | Structure of cyanobacterial photosystem I-IsiA supercomplex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Cao, P, Cao, D.F, Si, L, Su, X.D, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for energy and electron transfer of the photosystem I-IsiA-flavodoxin supercomplex.
Nat.Plants, 6, 2020
|
|
6OGZ
 
 | | In situ structure of Rotavirus RNA-dependent RNA polymerase at transcript-elongated state | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Inner capsid protein VP2, RNA (5'-R(P*AP*UP*AP*UP*AP*UP*AP*UP*AP*UP*AP*UP*AP*UP*AP*UP*A)-3'), ... | | Authors: | Ding, K, Chang, T, Shen, W, Roy, P, Zhou, Z.H. | | Deposit date: | 2019-04-03 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | In situ structures of rotavirus polymerase in action and mechanism of mRNA transcription and release.
Nat Commun, 10, 2019
|
|
6XRM
 
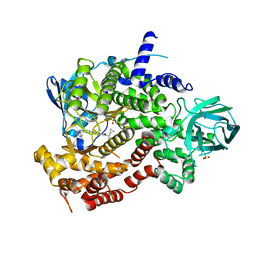 | | Crystal structure of human PI3K-gamma in complex with Compound 4 | | Descriptor: | 5-[2-amino-3-(1-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrimidin-5-yl]-2-[(1S)-1-cyclopropylethyl]-7-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Walker, N.P, Jeffrey, J.L. | | Deposit date: | 2020-07-13 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Discovery of Potent and Selective PI3K gamma Inhibitors.
J.Med.Chem., 63, 2020
|
|
6KE4
 
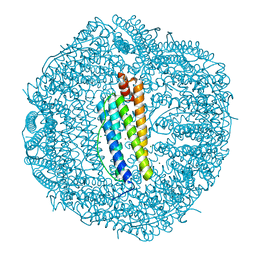 | | ABloop reengineered Ferritin Nanocage | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Wang, W.M, Wang, H.F. | | Deposit date: | 2019-07-03 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AB loop engineered ferritin nanocages for drug loading under benign experimental conditions.
Chem.Commun.(Camb.), 55, 2019
|
|
6XRN
 
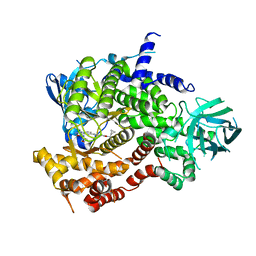 | | Crystal structure of human PI3K-gamma in complex with Compound 17 | | Descriptor: | 2-amino-5-{2-[(1S)-1-cyclopropylethyl]-7-methyl-1-oxo-2,3-dihydro-1H-isoindol-5-yl}-N-(trans-3-hydroxycyclobutyl)pyrazolo[1,5-a]pyrimidine-3-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Walker, N.P, Jeffrey, J.L. | | Deposit date: | 2020-07-13 | | Release date: | 2021-11-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationship Optimization of Pyrazolopyrimidine Amide Inhibitors of Phosphoinositide 3-Kinase gamma (PI3K gamma ).
J.Med.Chem., 65, 2022
|
|
6OGY
 
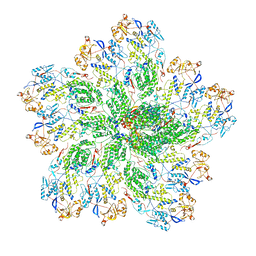 | | In situ structure of Rotavirus RNA-dependent RNA polymerase at duplex-open state | | Descriptor: | DNA/RNA (5'-D(*(GTG))-R(P*GP*C)-3'), Inner capsid protein VP2, RNA (5'-R(P*AP*GP*CP*C)-3'), ... | | Authors: | Ding, K, Chang, T, Shen, W, Roy, P, Zhou, Z.H. | | Deposit date: | 2019-04-03 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | In situ structures of rotavirus polymerase in action and mechanism of mRNA transcription and release.
Nat Commun, 10, 2019
|
|
6XRL
 
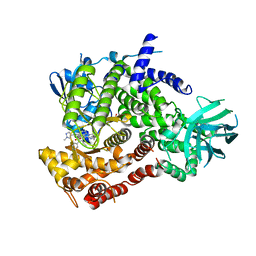 | | Crystal structure of human PI3K-gamma in complex with inhibitor IPI-549 | | Descriptor: | 2-amino-N-[(1S)-1-{8-[(1-methyl-1H-pyrazol-4-yl)ethynyl]-1-oxo-2-phenyl-1,2-dihydroisoquinolin-3-yl}ethyl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Walker, N.P, Jeffrey, J.L. | | Deposit date: | 2020-07-13 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Discovery of Potent and Selective PI3K gamma Inhibitors.
J.Med.Chem., 63, 2020
|
|
6BH3
 
 | |
6BGW
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(4,4-difluoropiperidin-1-yl)ethoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid(Compound N41) | | Descriptor: | 2-{(S)-(2-chlorophenyl)[2-(4,4-difluoropiperidin-1-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.644 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BGZ
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(1-methyl-1H-imidazol-2-yl)ethoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N47) | | Descriptor: | 2-{(S)-(2-chlorophenyl)[2-(1-methyl-1H-imidazol-2-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, Lysine-specific demethylase 5A, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.688 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BH1
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR (S)-2-((2-chlorophenyl)(2-(piperidin-1-yl)ethoxy)methyl)-1l2-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N52) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[2-(piperidin-1-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BGV
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(piperidin-1-yl)ethoxy)methyl)-1l2-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N40) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[2-(piperidin-1-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BH5
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(3-(piperidin-1-yl)propoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N48) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[3-(piperidin-1-yl)propoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
7N2O
 
 | | AS4.2-YEIH-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2Q
 
 | | AS4.3-YEIH-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AS4.3 T cell receptor alpha chain, AS4.3 T cell receptor beta chain, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2S
 
 | | AS3.1-PRPF3-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2R
 
 | | AS4.3-PRPF3-HLA*B27 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, AS4.3 T cell receptor alpha chain, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
