3A6V
 
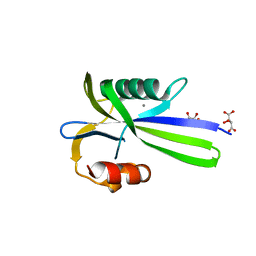 | |
3A6T
 
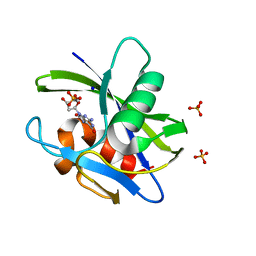 | | Crystal structure of MutT-8-OXO-DGMP complex | | Descriptor: | 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, Mutator mutT protein, SODIUM ION, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2009-09-09 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and dynamic features of the MutT protein in the recognition of nucleotides with the mutagenic 8-oxoguanine base
J.Biol.Chem., 285, 2010
|
|
3A6U
 
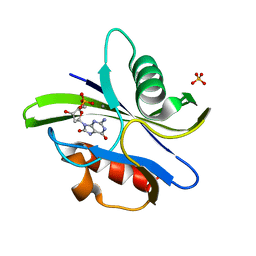 | | Crystal structure of MutT-8-OXO-dGMP-MN(II) complex | | Descriptor: | 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, Mutator mutT protein, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2009-09-09 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural and dynamic features of the MutT protein in the recognition of nucleotides with the mutagenic 8-oxoguanine base
J.Biol.Chem., 285, 2010
|
|
3A6S
 
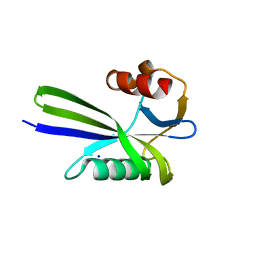 | | Crystal structure of the MutT protein | | Descriptor: | L(+)-TARTARIC ACID, Mutator mutT protein, SODIUM ION | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2009-09-09 | | Release date: | 2009-10-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and dynamic features of the MutT protein in the recognition of nucleotides with the mutagenic 8-oxoguanine base
J.Biol.Chem., 285, 2010
|
|
3A1M
 
 | | A fusion protein of a beta helix region of gene product 5 and the foldon region of bacteriophage T4 | | Descriptor: | POTASSIUM ION, chimera of thrombin cleavage site, Tail-associated lysozyme, ... | | Authors: | Yokoi, N, Suzuki, A, Hikage, T, Koshiyama, T, Terauchi, M, Yutani, K, Kanamaru, S, Arisaka, F, Yamane, T, Watanabe, Y, Ueno, T. | | Deposit date: | 2009-04-11 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Construction of Robust Bio-nanotubes using the Controlled Self-Assembly of Component Proteins of Bacteriophage T4
Small, 6, 2010
|
|
8EE2
 
 | |
8E1C
 
 | |
8E1B
 
 | |
8E2N
 
 | |
8EVD
 
 | |
8F6U
 
 | |
8F6V
 
 | |
8GJR
 
 | |
7COV
 
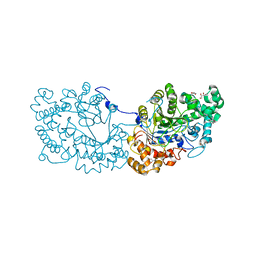 | | Potato D-enzyme, native (substrate free) | | Descriptor: | 4-alpha-glucanotransferase, chloroplastic/amyloplastic, CALCIUM ION, ... | | Authors: | Unno, H, Imamura, K. | | Deposit date: | 2020-08-05 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis and reaction mechanism of the disproportionating enzyme (D-enzyme) from potato.
Protein Sci., 29, 2020
|
|
6LX2
 
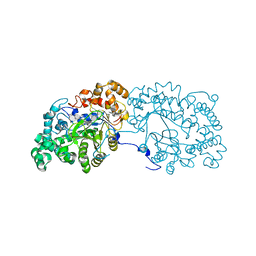 | | Potato D-enzyme complexed with CA26 | | Descriptor: | 4-alpha-glucanotransferase, chloroplastic/amyloplastic, 4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose, ... | | Authors: | Unno, H, Imamura, K. | | Deposit date: | 2020-02-10 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural analysis and reaction mechanism of the disproportionating enzyme (D-enzyme) from potato.
Protein Sci., 29, 2020
|
|
6LX1
 
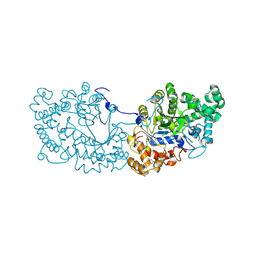 | | Potato D-enzyme complexed with Acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-1,5-anhydro-D-glucitol, 4-alpha-glucanotransferase, chloroplastic/amyloplastic, ... | | Authors: | Unno, H, Imamura, K. | | Deposit date: | 2020-02-10 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural analysis and reaction mechanism of the disproportionating enzyme (D-enzyme) from potato.
Protein Sci., 29, 2020
|
|
3VKN
 
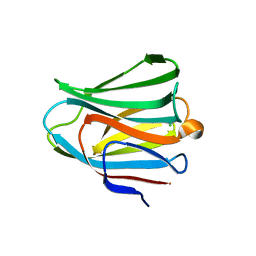 | | Galectin-8 N-terminal domain in free form | | Descriptor: | CHLORIDE ION, Galectin-8 | | Authors: | Kamitori, S, Yoshida, H. | | Deposit date: | 2011-11-18 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray structure of a protease-resistant mutant form of human galectin-8 with two carbohydrate recognition domains
Febs J., 279, 2012
|
|
1I1Z
 
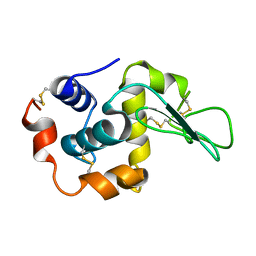 | | MUTANT HUMAN LYSOZYME (Q86D) | | Descriptor: | LYSOZYME C | | Authors: | Kuroki, R. | | Deposit date: | 2001-02-05 | | Release date: | 2001-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic responses of mutations at a Ca2+ binding site engineered into human lysozyme.
J.Biol.Chem., 273, 1998
|
|
1I20
 
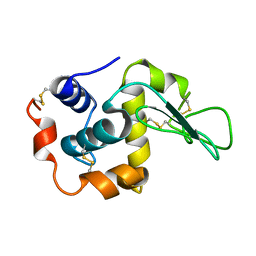 | | MUTANT HUMAN LYSOZYME (A92D) | | Descriptor: | LYSOZYME C | | Authors: | Kuroki, R. | | Deposit date: | 2001-02-05 | | Release date: | 2001-02-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic responses of mutations at a Ca2+ binding site engineered into human lysozyme.
J.Biol.Chem., 273, 1998
|
|
1I22
 
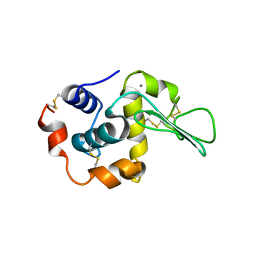 | | MUTANT HUMAN LYSOZYME (A83K/Q86D/A92D) | | Descriptor: | CALCIUM ION, LYSOZYME C | | Authors: | Kuroki, R. | | Deposit date: | 2001-02-05 | | Release date: | 2001-02-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic responses of mutations at a Ca2+ binding site engineered into human lysozyme.
J.Biol.Chem., 273, 1998
|
|
3VKL
 
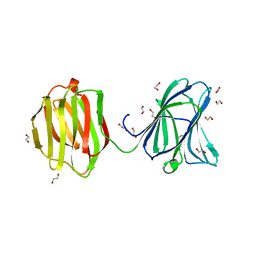 | |
3VKO
 
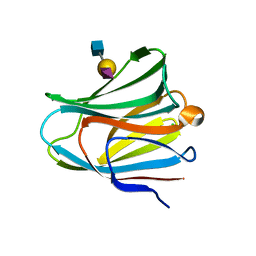 | | Galectin-8 N-terminal domain in complex with sialyllactosamine | | Descriptor: | CHLORIDE ION, Galectin-8, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kamitori, S, Yoshida, H. | | Deposit date: | 2011-11-18 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | X-ray structure of a protease-resistant mutant form of human galectin-8 with two carbohydrate recognition domains
Febs J., 279, 2012
|
|
2ZU0
 
 | |
3WNO
 
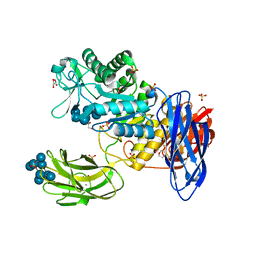 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with cycloisomaltooctaose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
3WUS
 
 | | Crystal Structure of the Vif-Binding Domain of Human APOBEC3F | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3F, ZINC ION | | Authors: | Nakashima, M, Kawamura, T, Ode, H, Watanabe, N, Iwatani, Y. | | Deposit date: | 2014-05-02 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural Insights into HIV-1 Vif-APOBEC3F Interaction.
J.Virol., 90, 2015
|
|
