5VF5
 
 | | Crystal structure of the vicilin from Solanum melongena, re-refinement | | Descriptor: | ACETATE ION, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Porebski, P.J, Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VLB
 
 | |
6B92
 
 | | Crystal Structure of the N-terminal domain of human METTL16 in complex with SAH | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, U6 small nuclear RNA (adenine-(43)-N(6))-methyltransferase | | Authors: | Ruszkowska, A, Ruszkowski, M, Dauter, Z, Brown, J.A. | | Deposit date: | 2017-10-09 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the RNA methyltransferase domain of METTL16.
Sci Rep, 8, 2018
|
|
6BQ7
 
 | |
6BQ2
 
 | |
6CZY
 
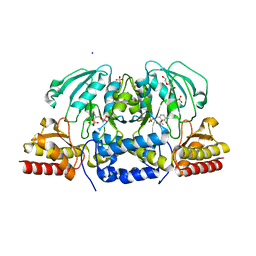 | | Crystal structure of Arabidopsis thaliana phosphoserine aminotransferase isoform 1 (AtPSAT1) in complex with Pyridoxamine-5'-phosphate (PMP) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2018-04-09 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Analysis of Phosphoserine Aminotransferase (Isoform 1) FromArabidopsis thaliana- the Enzyme Involved in the Phosphorylated Pathway of Serine Biosynthesis.
Front Plant Sci, 9, 2018
|
|
6CD0
 
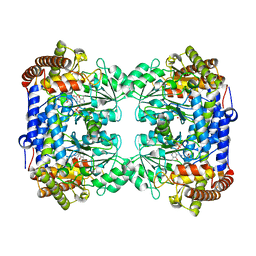 | | Crystal structure of Medicago truncatula serine hydroxymethyltransferase 3 (MtSHMT3), PLP-internal aldimine and apo form | | Descriptor: | ACETATE ION, FORMIC ACID, Serine hydroxymethyltransferase | | Authors: | Ruszkowski, M, Sekula, B, Ruszkowska, A, Dauter, Z. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Chloroplastic Serine Hydroxymethyltransferase FromMedicago truncatula: A Structural Characterization.
Front Plant Sci, 9, 2018
|
|
6CZX
 
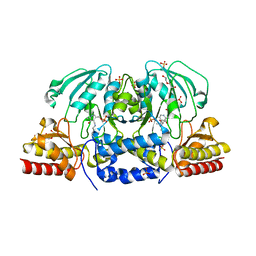 | | Crystal structure of Arabidopsis thaliana phosphoserine aminotransferase isoform 1 (AtPSAT1) in complex with PLP internal aldimine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, DI(HYDROXYETHYL)ETHER, Phosphoserine aminotransferase 1, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2018-04-09 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Analysis of Phosphoserine Aminotransferase (Isoform 1) FromArabidopsis thaliana- the Enzyme Involved in the Phosphorylated Pathway of Serine Biosynthesis.
Front Plant Sci, 9, 2018
|
|
6CZZ
 
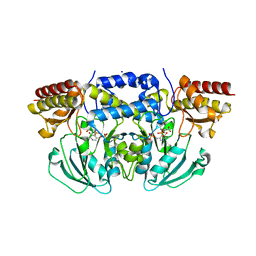 | | Crystal structure of Arabidopsis thaliana phosphoserine aminotransferase isoform 1 (AtPSAT1) in complex with PLP-phosphoserine geminal diamine intermediate | | Descriptor: | PHOSPHOSERINE, PYRIDOXAL-5'-PHOSPHATE, Phosphoserine aminotransferase 1, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2018-04-09 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis of Phosphoserine Aminotransferase (Isoform 1) FromArabidopsis thaliana- the Enzyme Involved in the Phosphorylated Pathway of Serine Biosynthesis.
Front Plant Sci, 9, 2018
|
|
3LJC
 
 | | Crystal structure of Lon N-terminal domain. | | Descriptor: | ATP-dependent protease La | | Authors: | Li, M, Gustchina, A, Dauter, Z, Wlodawer, A. | | Deposit date: | 2010-01-26 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the N-terminal fragment of Escherichia coli Lon protease
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6BQ5
 
 | | Crystal structure of Medicago truncatula Thermospermine Synthase (MtTSPS) in complex with 5'-methylthioadenosine | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, GLYCEROL, ... | | Authors: | Sekula, B, Dauter, Z. | | Deposit date: | 2017-11-27 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of thermospermine synthase fromMedicago truncatulaand substrate discriminatory features of plant aminopropyltransferases.
Biochem. J., 475, 2018
|
|
6BQ4
 
 | | Crystal structure of Medicago truncatula Thermospermine Synthase (MtTSPS) in complex with adenosine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE, ... | | Authors: | Sekula, B, Dauter, Z. | | Deposit date: | 2017-11-27 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of thermospermine synthase fromMedicago truncatulaand substrate discriminatory features of plant aminopropyltransferases.
Biochem. J., 475, 2018
|
|
3LF3
 
 | | Crystal Structure of Fast Fluorescent Timer Fast-FT | | Descriptor: | Fast Fluorescent Timer Fast-FT | | Authors: | Pletnev, S, Dauter, Z. | | Deposit date: | 2010-01-15 | | Release date: | 2010-03-09 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Understanding blue-to-red conversion in monomeric fluorescent timers and hydrolytic degradation of their chromophores
J.Am.Chem.Soc., 132, 2010
|
|
6CCZ
 
 | | Crystal structure of Medicago truncatula serine hydroxymethyltransferase 3 (MtSHMT3) soaked with selenourea | | Descriptor: | ACETATE ION, FORMIC ACID, Serine hydroxymethyltransferase, ... | | Authors: | Ruszkowski, M, Sekula, B, Ruszkowska, A, Dauter, Z. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-23 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Chloroplastic Serine Hydroxymethyltransferase FromMedicago truncatula: A Structural Characterization.
Front Plant Sci, 9, 2018
|
|
6BQ3
 
 | |
3LF4
 
 | | Crystal Structure of Fluorescent Timer Precursor Blue102 | | Descriptor: | Fluorescent Timer Precursor Blue102 | | Authors: | Pletnev, S, Dauter, Z. | | Deposit date: | 2010-01-15 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.808 Å) | | Cite: | Understanding blue-to-red conversion in monomeric fluorescent timers and hydrolytic degradation of their chromophores
J.Am.Chem.Soc., 132, 2010
|
|
6BQ6
 
 | |
3LZT
 
 | | REFINEMENT OF TRICLINIC LYSOZYME AT ATOMIC RESOLUTION | | Descriptor: | ACETATE ION, LYSOZYME, NITRATE ION | | Authors: | Walsh, M.A, Schneider, T, Sieker, L.C, Dauter, Z, Lamzin, V, Wilson, K.S. | | Deposit date: | 1997-03-23 | | Release date: | 1998-03-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (0.925 Å) | | Cite: | Refinement of triclinic hen egg-white lysozyme at atomic resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
6CD1
 
 | | Crystal structure of Medicago truncatula serine hydroxymethyltransferase 3 (MtSHMT3), complexes with reaction intermediates | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCINE, ... | | Authors: | Ruszkowski, M, Sekula, B, Ruszkowska, A, Dauter, Z. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Chloroplastic Serine Hydroxymethyltransferase FromMedicago truncatula: A Structural Characterization.
Front Plant Sci, 9, 2018
|
|
3NR6
 
 | | Crystal structure of xenotropic murine leukemia virus-related virus (XMRV) protease | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Protease p14 | | Authors: | Lubkowski, J, Li, M, Gustchina, A, Zhou, D, Dauter, Z, Wlodawer, A. | | Deposit date: | 2010-06-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of XMRV protease differs from the structures of other retropepsins.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1X3E
 
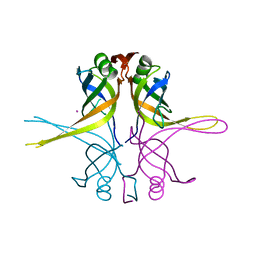 | | Crystal structure of the single-stranded DNA-binding protein from Mycobacterium smegmatis | | Descriptor: | CADMIUM ION, Single-strand binding protein | | Authors: | Saikrishnan, K, Manjunath, G.P, Singh, P, Jeyakanthan, J, Dauter, Z, Sekar, K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2005-05-04 | | Release date: | 2005-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologus SSBs: biological implications of structural plasticity and variability in quaternary association.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1X3G
 
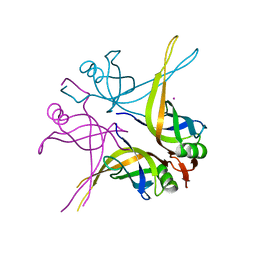 | | Crystal structure of the single-stranded DNA-binding protein from Mycobacterium SMEGMATIS | | Descriptor: | CADMIUM ION, Single-strand binding protein | | Authors: | Saikrishnan, K, Manjunath, G.P, Singh, P, Jeyakanthan, J, Dauter, Z, Sekar, K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2005-05-05 | | Release date: | 2005-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologus SSBs: biological implications of structural plasticity and variability in quaternary association.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1WQ6
 
 | | The tetramer structure of the nervy homolgy two (NHR2) domain of AML1-ETO is critical for AML1-ETO'S activity | | Descriptor: | AML1-ETO | | Authors: | Liu, Y, Cheney, M.D, Chruszcz, M, Lukasik, S.M, Hartman, K.L, Laue, T.M, Dauter, Z, Minor, W, Speck, N.A, Bushweller, J.H. | | Deposit date: | 2004-09-23 | | Release date: | 2005-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO's activity
Cancer Cell, 9, 2006
|
|
1X3F
 
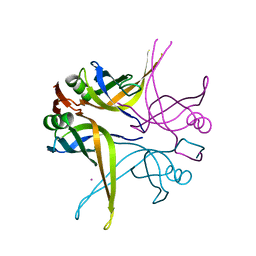 | | Crystal structure of the single-stranded DNA-binding protein from Mycobacterium SMEGMATIS | | Descriptor: | CADMIUM ION, Single-strand binding protein | | Authors: | Saikrishnan, K, Manjunath, G.P, Singh, P, Jeyakanthan, J, Dauter, Z, Sekar, K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2005-05-05 | | Release date: | 2005-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologus SSBs: biological implications of structural plasticity and variability in quaternary association.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1W9X
 
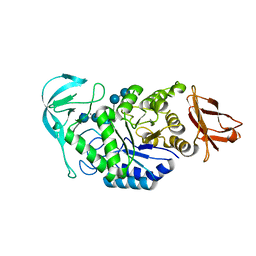 | | Bacillus halmapalus alpha amylase | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ALPHA AMYLASE, CALCIUM ION, ... | | Authors: | Davies, G.J, Brzozowski, A.M, Dauter, Z, Rasmussen, M.D, Borchert, T.V, Wilson, K.S. | | Deposit date: | 2004-10-20 | | Release date: | 2005-02-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a Bacillus Halmapalus Family 13 Alpha-Amylase, Bha, in Complex with an Acarbose-Derived Nonasaccharide at 2.1 A Resolution
Acta Crystallogr.,Sect.D, 61, 2005
|
|
