7O5H
 
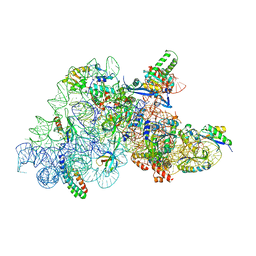 | | Ribosomal methyltransferase KsgA bound to small ribosomal subunit | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Stephan, N.C, Ries, A.B, Boehringer, D, Ban, N. | | Deposit date: | 2021-04-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of successive adenosine modifications by the conserved ribosomal methyltransferase KsgA.
Nucleic Acids Res., 49, 2021
|
|
7ODS
 
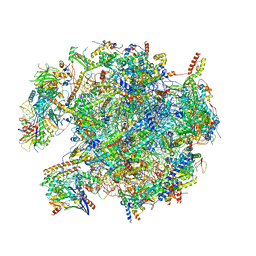 | | State B of the human mitoribosomal large subunit assembly intermediate | | Descriptor: | 16S mitochondrial rRNA, DNA (30-MER),16S mitochondrial rRNA, 39S ribosomal protein L10, ... | | Authors: | Lenarcic, T, Jaskolowski, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Saurer, M, Lee, R.G, Rackham, O, Filipovska, A, Ban, N. | | Deposit date: | 2021-04-30 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Stepwise maturation of the peptidyl transferase region of human mitoribosomes.
Nat Commun, 12, 2021
|
|
7ODT
 
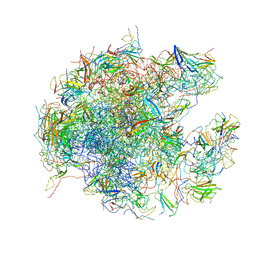 | | State C of the human mitoribosomal large subunit assembly intermediate | | Descriptor: | 16S mitochondrial rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Lenarcic, T, Jaskolowski, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Saurer, M, Lee, R.G, Rackham, O, Filipovska, A, Ban, N. | | Deposit date: | 2021-04-30 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Stepwise maturation of the peptidyl transferase region of human mitoribosomes.
Nat Commun, 12, 2021
|
|
7ODR
 
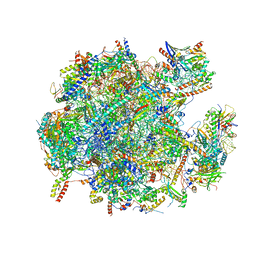 | | State A of the human mitoribosomal large subunit assembly intermediate | | Descriptor: | 16S mitochondrial rRNA, DNA (31-MER),16S mitochondrial rRNA, 39S ribosomal protein L10, ... | | Authors: | Lenarcic, T, Jaskolowski, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Saurer, M, Lee, R.G, Rackham, O, Filipovska, A, Ban, N. | | Deposit date: | 2021-04-30 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Stepwise maturation of the peptidyl transferase region of human mitoribosomes.
Nat Commun, 12, 2021
|
|
4C7O
 
 | | The structural basis of FtsY recruitment and GTPase activation by SRP RNA | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SIGNAL RECOGNITION PARTICLE PROTEIN, ... | | Authors: | Voigts-Hoffmann, F, Schmitz, N, Shen, K, Shan, S.O, Ataide, S.F, Ban, N. | | Deposit date: | 2013-09-23 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structural Basis of Ftsy Recruitment and Gtpase Activation by Srp RNA
Mol.Cell, 52, 2013
|
|
3D2V
 
 | |
3D2X
 
 | | Structure of the thiamine pyrophosphate-specific riboswitch bound to oxythiamine pyrophosphate | | Descriptor: | 3-[(4-hydroxy-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, MAGNESIUM ION, TPP-specific riboswitch | | Authors: | Thore, S, Frick, C, Ban, N. | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of thiamine pyrophosphate analogues binding to the eukaryotic riboswitch
J.Am.Chem.Soc., 130, 2008
|
|
1ULG
 
 | | CGL2 in complex with Thomsen-Friedenreich antigen | | Descriptor: | beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1W26
 
 | | Trigger Factor in Complex with the Ribosome forms a Molecular Cradle for Nascent Proteins | | Descriptor: | TRIGGER FACTOR | | Authors: | Ferbitz, L, Maier, T, Patzelt, H, Bukau, B, Deuerling, E, Ban, N. | | Deposit date: | 2004-06-28 | | Release date: | 2004-09-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Trigger Factor in Complex with the Ribosome Forms a Molecular Cradle for Nascent Proteins
Nature, 431, 2004
|
|
1W2B
 
 | | Trigger Factor ribosome binding domain in complex with 50S | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L13P, ... | | Authors: | Ferbitz, L, Maier, T, Patzelt, H, Bukau, B, Deuerling, E, Ban, N. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Trigger Factor in Complex with the Ribosome Forms a Molecular Cradle for Nascent Proteins
Nature, 431, 2004
|
|
1UL9
 
 | | CGL2 ligandfree | | Descriptor: | galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ULC
 
 | | CGL2 in complex with lactose | | Descriptor: | beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ULF
 
 | | CGL2 in complex with Blood Group A tetrasaccharide | | Descriptor: | alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ULE
 
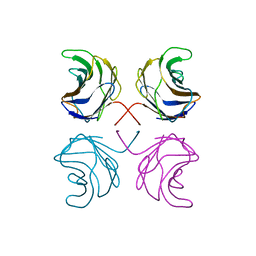 | | CGL2 in complex with linear B2 trisaccharide | | Descriptor: | alpha-D-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ULD
 
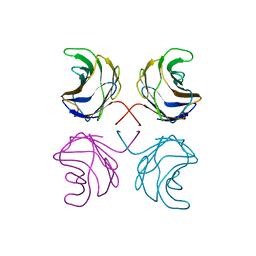 | | CGL2 in complex with blood group H type II | | Descriptor: | alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
4ADV
 
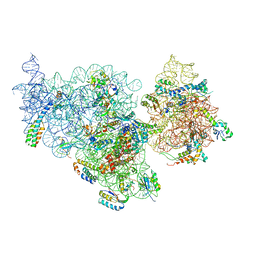 | | Structure of the E. coli methyltransferase KsgA bound to the E. coli 30S ribosomal subunit | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Boehringer, D, O'Farrell, H.C, Rife, J.P, Ban, N. | | Deposit date: | 2012-01-03 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Structural Insights Into Methyltransferase Ksga Function in 30S Ribosomal Subunit Biogenesis
J.Biol.Chem., 287, 2012
|
|
4AC9
 
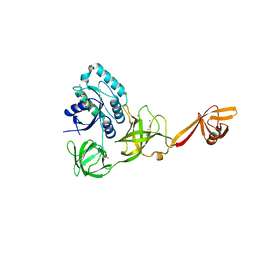 | | CRYSTAL STRUCTURE OF TRANSLATION ELONGATION FACTOR SELB FROM METHANOCOCCUS MARIPALUDIS IN COMPLEX WITH GDP | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Leibundgut, M, Frick, C, Thanbichler, M, Boeck, A, Ban, N. | | Deposit date: | 2011-12-14 | | Release date: | 2012-08-22 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Selenocysteine tRNA-Specific Elongation Factor Selb is a Structural Chimaera of Elongation and Initiation Factors.
Embo J., 24, 2005
|
|
4A29
 
 | | Structure of the engineered retro-aldolase RA95.0 | | Descriptor: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, D-MALATE, ENGINEERED RETRO-ALDOL ENZYME RA95.0 | | Authors: | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | Deposit date: | 2011-09-23 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
4A2R
 
 | | Structure of the engineered retro-aldolase RA95.5-5 | | Descriptor: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE | | Authors: | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | Deposit date: | 2011-09-28 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
4ACA
 
 | | CRYSTAL STRUCTURE OF TRANSLATION ELONGATION FACTOR SELB FROM METHANOCOCCUS MARIPALUDIS, APO FORM | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GUANOSINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Leibundgut, M, Frick, C, Thanbichler, M, Boeck, A, Ban, N. | | Deposit date: | 2011-12-14 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Selenocysteine tRNA-Specific Elongation Factor Selb is a Structural Chimaera of Elongation and Initiation Factors.
Embo J., 24, 2005
|
|
4ACB
 
 | | CRYSTAL STRUCTURE OF TRANSLATION ELONGATION FACTOR SELB FROM METHANOCOCCUS MARIPALUDIS IN COMPLEX WITH THE GTP ANALOGUE GPPNHP | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Leibundgut, M, Frick, C, Thanbichler, M, Boeck, A, Ban, N. | | Deposit date: | 2011-12-14 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Selenocysteine tRNA-Specific Elongation Factor Selb is a Structural Chimaera of Elongation and Initiation Factors.
Embo J., 24, 2005
|
|
3J8B
 
 | | Model of the human eIF3 PCI-MPN octamer docked into the 43S-HCV IRES EM map | | Descriptor: | Eukaryotic translation initiation factor 3 subunit A, Eukaryotic translation initiation factor 3 subunit C, Eukaryotic translation initiation factor 3 subunit E, ... | | Authors: | Erzberger, J.P, Ban, N. | | Deposit date: | 2014-10-08 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex.
Cell(Cambridge,Mass.), 158, 2014
|
|
3J8C
 
 | | Model of the human eIF3 PCI-MPN octamer docked into the 43S EM map | | Descriptor: | Eukaryotic translation initiation factor 3 subunit A, Eukaryotic translation initiation factor 3 subunit C, Eukaryotic translation initiation factor 3 subunit E, ... | | Authors: | Erzberger, J.P, Ban, N. | | Deposit date: | 2014-10-08 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.6 Å) | | Cite: | Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex.
Cell(Cambridge,Mass.), 158, 2014
|
|
4ADX
 
 | | The Cryo-EM Structure of the Archaeal 50S Ribosomal Subunit in Complex with Initiation Factor 6 | | Descriptor: | 23S Ribosomal RNA EXPANSION SEGMENTS, 23S ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Greber, B.J, Boehringer, D, Godinic-Mikulcic, V, Crnkovic, A, Ibba, M, Weygand-Durasevic, I, Ban, N. | | Deposit date: | 2012-01-04 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-Em Structure of the Archaeal 50S Ribosomal Subunit in Complex with Initiation Factor 6 and Implications for Ribosome Evolution
J.Mol.Biol., 418, 2012
|
|
4A2S
 
 | | Structure of the engineered retro-aldolase RA95.5 | | Descriptor: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE | | Authors: | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | Deposit date: | 2011-09-28 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
