1RFS
 
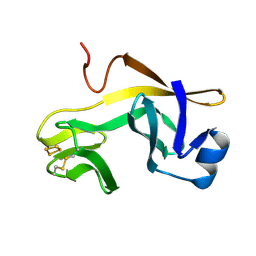 | | RIESKE SOLUBLE FRAGMENT FROM SPINACH | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, RIESKE PROTEIN | | Authors: | Carrell, C.J, Zhang, H, Cramer, W.A, Smith, J.L. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Biological identity and diversity in photosynthesis and respiration: structure of the lumen-side domain of the chloroplast Rieske protein.
Structure, 5, 1997
|
|
4GOX
 
 | |
4GBM
 
 | |
4H5O
 
 | |
4H5Q
 
 | |
4H5L
 
 | |
1CI3
 
 | | CYTOCHROME F FROM THE B6F COMPLEX OF PHORMIDIUM LAMINOSUM | | Descriptor: | HEME C, PROTEIN (CYTOCHROME F), ZINC ION | | Authors: | Carrell, C.J, Schlarb, B.G, Howe, C.J, Bendall, D.S, Cramer, W.A, Smith, J.L. | | Deposit date: | 1999-04-07 | | Release date: | 1999-08-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the soluble domain of cytochrome f from the cyanobacterium Phormidium laminosum.
Biochemistry, 38, 1999
|
|
9E56
 
 | |
9E4S
 
 | |
9E4U
 
 | |
9E4X
 
 | |
4H5M
 
 | |
4H5P
 
 | |
9EBZ
 
 | |
1CTM
 
 | | CRYSTAL STRUCTURE OF CHLOROPLAST CYTOCHROME F REVEALS A NOVEL CYTOCHROME FOLD AND UNEXPECTED HEME LIGATION | | Descriptor: | CYTOCHROME F, HEME C | | Authors: | Martinez, S.E, Huang, D, Szczepaniak, A, Cramer, W.A, Smith, J.L. | | Deposit date: | 1994-01-02 | | Release date: | 1994-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of chloroplast cytochrome f reveals a novel cytochrome fold and unexpected heme ligation.
Structure, 2, 1994
|
|
4HXY
 
 | |
9CGL
 
 | | Pikromycin Thioesterase Doubly Protected DAP | | Descriptor: | 2-{[(1R)-1-(6-nitro-2H-1,3-benzodioxol-5-yl)ethyl]sulfanyl}ethyl formate, Narbonolide/10-deoxymethynolide synthase PikA4, module 6 | | Authors: | McCullough, T.M, Smith, J.L. | | Deposit date: | 2024-06-29 | | Release date: | 2024-09-18 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Substrate Trapping in Polyketide Synthase Thioesterase Domains: Structural Basis for Macrolactone Formation.
Acs Catalysis, 14, 2024
|
|
9CBD
 
 | | Pikromycin Thioesterase Domain | | Descriptor: | Narbonolide/10-deoxymethynolide synthase PikA4, module 6 | | Authors: | McCullough, T.M, Smith, J.L. | | Deposit date: | 2024-06-19 | | Release date: | 2024-09-18 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Trapping in Polyketide Synthase Thioesterase Domains: Structural Basis for Macrolactone Formation.
Acs Catalysis, 14, 2024
|
|
9CFJ
 
 | | Fluvirucin Thioesterase Domain (FluC TE) | | Descriptor: | FluC, GLYCEROL, PENTAETHYLENE GLYCOL | | Authors: | Choudhary, V, Smith, J.L. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-18 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Substrate Trapping in Polyketide Synthase Thioesterase Domains: Structural Basis for Macrolactone Formation.
Acs Catalysis, 14, 2024
|
|
9EAT
 
 | |
9EAW
 
 | |
6MBH
 
 | |
6MFD
 
 | | GphF GNAT-like decarboxylase in complex with isobutyryl-CoA | | Descriptor: | ACETATE ION, GLYCEROL, GphF, ... | | Authors: | Skiba, M.A, Tran, C.L, Smith, J.L. | | Deposit date: | 2018-09-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Repurposing the GNAT Fold in the Initiation of Polyketide Biosynthesis.
Structure, 28, 2020
|
|
6MFC
 
 | | GphF GNAT-like decarboxylase | | Descriptor: | GLYCEROL, GphF, PENTAETHYLENE GLYCOL | | Authors: | Skiba, M.A, Tran, C.L, Smith, J.L. | | Deposit date: | 2018-09-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.589 Å) | | Cite: | Repurposing the GNAT Fold in the Initiation of Polyketide Biosynthesis.
Structure, 28, 2020
|
|
8SY3
 
 | |
