6P58
 
 | | Dark and Steady State-Illuminated Crystal Structure of Cyanobacteriochrome Receptor PixJ at 150K | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Clinger, J.A, Miller, M.D, Buirgie, E.S, Vierstra, R.D, Phillips Jr, G.N. | | Deposit date: | 2019-05-29 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PRU
 
 | | Photoconvertible crystals of PixJ from Thermosynechococcus elongatus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Burgie, E.S, Clinger, J.A, Miller, M.D, Phillips Jr, G.N, Vierstra, R.D. | | Deposit date: | 2019-07-11 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8BT3
 
 | |
8BT4
 
 | | Ribonucleotide Reductase class Ie R2 from Mesoplasma florum, radical-lost ground state | | Descriptor: | CALCIUM ION, GLYCEROL, Ribonucleoside-diphosphate reductase | | Authors: | Lebrette, H, Srinivas, V, Hogbom, M. | | Deposit date: | 2022-11-27 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of a ribonucleotide reductase R2 protein radical.
Science, 382, 2023
|
|
3LR7
 
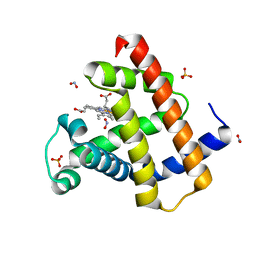 | | Ferric horse heart myoglobin, nitrite adduct | | Descriptor: | Myoglobin, NITRITE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yi, J, Richter-Addo, G.B. | | Deposit date: | 2010-02-10 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Synchrotron X-ray-Induced Photoreduction of Ferric Myoglobin Nitrite Crystals Gives the Ferrous Derivative with Retention of the O-Bonded Nitrite Ligand.
Biochemistry, 49, 2010
|
|
6GUZ
 
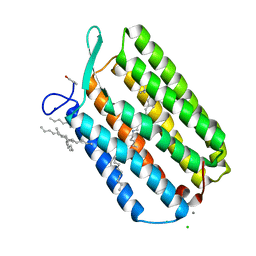 | | Ground state structure of Archaerhodopsin-3 obtained from LCP crystals using a thin-film sandwich at room temperature | | Descriptor: | Archaerhodopsin-3, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Moraes, I, Judge, P.J, Bada Juarez, J.F, Vinals, J, Axford, D, Watts, A. | | Deposit date: | 2018-06-19 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Room temperature structure of the Archaerhodopsin-3 obtained from LCP crystals using a thin-film sandwich
To Be Published
|
|
6GUY
 
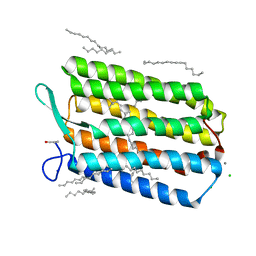 | | Room temperature structure of Archaerhodopsin-3 via LCP extruder using synchrotron radiation | | Descriptor: | Archaerhodopsin-3, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Moraes, I, Judge, P.J, Axford, D, Bada Juarez, J.F, Vinals, J, Watts, A. | | Deposit date: | 2018-06-19 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Room temperature structure of Archaerhodopsin-3 via LCP extruder using synchrotron radiation
To Be Published
|
|
5KAF
 
 | | RT XFEL structure of Photosystem II in the dark state at 3.0 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.00001 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5KAI
 
 | | NH3-bound RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 2.8 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.80000925 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
1DLQ
 
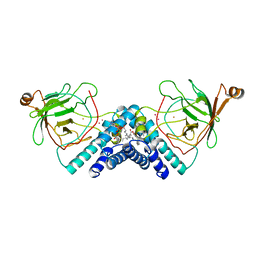 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER SP. ADP1 INHIBITED BY BOUND MERCURY | | Descriptor: | CATECHOL 1,2-DIOXYGENASE, FE (III) ION, MERCURY (II) ION, ... | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-11 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
1EO2
 
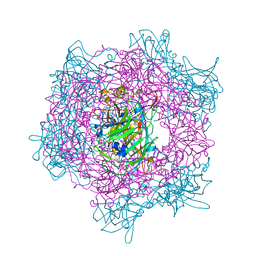 | | CRYSTAL STRUCTURE OF ACINETOBACTER SP. ADP1 PROTOCATECHUATE 3,4-DIOXYGENASE | | Descriptor: | FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, PROTOCATECHUATE 3,4-DIOXYGENASE BETA CHAIN | | Authors: | Vetting, M.W, D'Argenio, D.A, Ornston, L.N, Ohlendorf, D.H. | | Deposit date: | 2000-03-21 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Acinetobacter strain ADP1 protocatechuate 3, 4-dioxygenase at 2.2 A resolution: implications for the mechanism of an intradiol dioxygenase.
Biochemistry, 39, 2000
|
|
1DLT
 
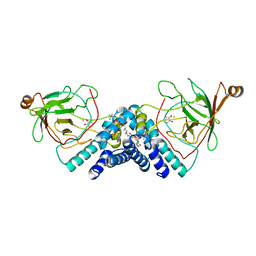 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER SP. ADP1 WITH BOUND CATECHOL | | Descriptor: | CATECHOL, CATECHOL 1,2-DIOXYGENASE, FE (III) ION, ... | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-12 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
1EO9
 
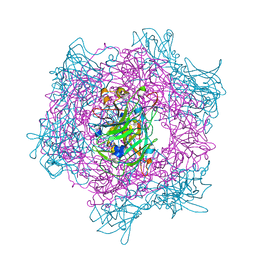 | | CRYSTAL STRUCTURE OF ACINETOBACTER SP. ADP1 PROTOCATECHUATE 3,4-DIOXYGENASE AT PH < 7.0 | | Descriptor: | FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, PROTOCATECHUATE 3,4-DIOXYGENASE BETA CHAIN;, ... | | Authors: | Vetting, M.W, D'Argenio, D.A, Ornston, L.N, Ohlendorf, D.H. | | Deposit date: | 2000-03-22 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Acinetobacter strain ADP1 protocatechuate 3, 4-dioxygenase at 2.2 A resolution: implications for the mechanism of an intradiol dioxygenase.
Biochemistry, 39, 2000
|
|
1EOB
 
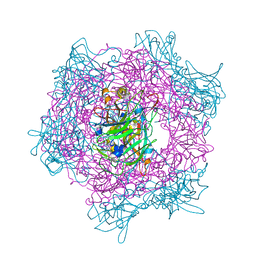 | | CRYSTAL STRUCTURE OF ACINETOBACTER SP. ADP1 PROTOCATECHUATE 3,4-DIOXYGENASE IN COMPLEX WITH 3,4-DIHYDROXYBENZOATE | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, D'Argenio, D.A, Ornston, L.N, Ohlendorf, D.H. | | Deposit date: | 2000-03-22 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Acinetobacter strain ADP1 protocatechuate 3, 4-dioxygenase at 2.2 A resolution: implications for the mechanism of an intradiol dioxygenase.
Biochemistry, 39, 2000
|
|
1EOC
 
 | | CRYSTAL STRUCTURE OF ACINETOBACTER SP. ADP1 PROTOCATECHUATE 3,4-DIOXYGENASE IN COMPLEX WITH 4-NITROCATECHOL | | Descriptor: | 4-NITROCATECHOL, FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, D'Argenio, D.A, Ornston, L.N, Ohlendorf, D.H. | | Deposit date: | 2000-03-22 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Acinetobacter strain ADP1 protocatechuate 3, 4-dioxygenase at 2.2 A resolution: implications for the mechanism of an intradiol dioxygenase.
Biochemistry, 39, 2000
|
|
1DMH
 
 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER SP. ADP1 WITH BOUND 4-METHYLCATECHOL | | Descriptor: | 4-METHYLCATECHOL, CATECHOL 1,2-DIOXYGENASE, FE (III) ION, ... | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
1DLM
 
 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER CALCOACETICUS NATIVE DATA | | Descriptor: | CATECHOL 1,2-DIOXYGENASE, FE (III) ION, [1-PENTADECANOYL-2-DECANOYL-GLYCEROL-3-YL]PHOSPHONYL CHOLINE | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-11 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
1EOA
 
 | | CRYSTAL STRUCTURE OF ACINETOBACTER SP. ADP1 PROTOCATECHUATE 3,4-DIOXYGENASE IN COMPLEX WITH CYANIDE | | Descriptor: | CYANIDE ION, FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, D'Argenio, D.A, Ornston, L.N, Ohlendorf, D.H. | | Deposit date: | 2000-03-22 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Acinetobacter strain ADP1 protocatechuate 3, 4-dioxygenase at 2.2 A resolution: implications for the mechanism of an intradiol dioxygenase.
Biochemistry, 39, 2000
|
|
4I25
 
 | | 2.00 Angstroms X-ray crystal structure of NAD- and substrate-bound 2-aminomuconate 6-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | (2E,4E)-2-amino-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huo, L, Davis, I, Chen, L, Liu, A. | | Deposit date: | 2012-11-21 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
4I26
 
 | | 2.20 Angstroms X-ray crystal structure of 2-aminomuconate 6-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | 1,2-ETHANEDIOL, 2-aminomuconate 6-semialdehyde dehydrogenase, SODIUM ION | | Authors: | Davis, I, Huo, L, Chen, L, Liu, A. | | Deposit date: | 2012-11-21 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
4I2R
 
 | | 2.15 Angstroms X-ray crystal structure of NAD- and alternative substrate-bound 2-aminomuconate 6-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | (2E,4E)-2-hydroxy-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huo, L, Davis, I, Chen, L, Liu, A. | | Deposit date: | 2012-11-22 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
4I1W
 
 | | 2.00 Angstroms X-ray crystal structure of NAD- bound 2-aminomuconate 6-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | 2-aminomuconate 6-semialdehyde dehydrogenase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Huo, L, Davis, I, Chen, L, Liu, A. | | Deposit date: | 2012-11-21 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
6WEB
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
6WEC
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
4NPI
 
 | | 1.94 Angstroms X-ray crystal structure of NAD- and intermediate- bound alpha-aminomuconate-epsilon-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | (2Z,4E)-2-hydroxy-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huo, L, Davis, I, Liu, F, Iwaki, H, Hasegawa, Y, Liu, A. | | Deposit date: | 2013-11-21 | | Release date: | 2014-12-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
