1G1W
 
 | | T4 LYSOZYME MUTANT C54T/C97A/Q105M | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Quillin, M.L, Matthews, B.W. | | Deposit date: | 2000-10-13 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
1FVA
 
 | | CRYSTAL STRUCTURE OF BOVINE METHIONINE SULFOXIDE REDUCTASE | | Descriptor: | PEPTIDE METHIONINE SULFOXIDE REDUCTASE | | Authors: | Lowther, W.T, Brot, N, Weissbach, H, Matthews, B.W. | | Deposit date: | 2000-09-19 | | Release date: | 2000-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and mechanism of peptide methionine sulfoxide reductase, an "anti-oxidation" enzyme.
Biochemistry, 39, 2000
|
|
1GCT
 
 | |
1HO8
 
 | |
1T8A
 
 | | USE OF SEQUENCE DUPLICATION TO ENGINEER A LIGAND-TRIGGERED LONG-DISTANCE MOLECULAR SWITCH IN T4 Lysozyme | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, GUANIDINE, ... | | Authors: | Yousef, M.S, Baase, W.A, Matthews, B.W. | | Deposit date: | 2004-05-11 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Use of sequence duplication to engineer a ligand-triggered, long-distance molecular switch in T4 lysozyme.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1T97
 
 | |
3F8V
 
 | |
3FA0
 
 | |
3FAD
 
 | |
1LNE
 
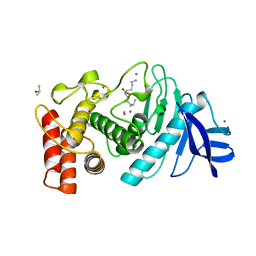 | | A STRUCTURAL ANALYSIS OF METAL SUBSTITUTIONS IN THERMOLYSIN | | Descriptor: | CADMIUM ION, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Holland, D.R, Hausrath, A.C, Juers, D, Matthews, B.W. | | Deposit date: | 1994-05-13 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of zinc substitutions in the active site of thermolysin.
Protein Sci., 4, 1995
|
|
1LNF
 
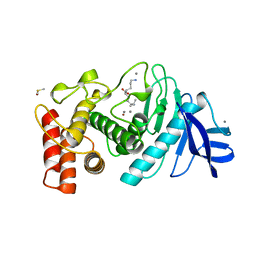 | |
1LND
 
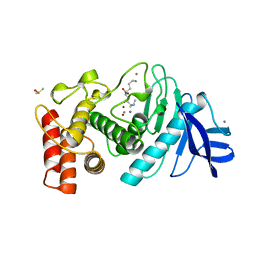 | | A STRUCTURAL ANALYSIS OF METAL SUBSTITUTIONS IN THERMOLYSIN | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, LYSINE, ... | | Authors: | Holland, D.R, Hausrath, A.C, Juers, D, Matthews, B.W. | | Deposit date: | 1994-05-13 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of zinc substitutions in the active site of thermolysin.
Protein Sci., 4, 1995
|
|
1LNB
 
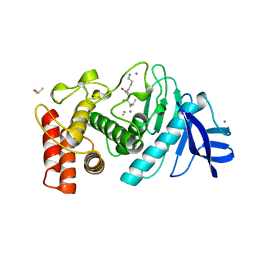 | | A STRUCTURAL ANALYSIS OF METAL SUBSTITUTIONS IN THERMOLYSIN | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, FE (III) ION, ... | | Authors: | Holland, D.R, Hausrath, A.C, Juers, D, Matthews, B.W. | | Deposit date: | 1994-05-13 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of zinc substitutions in the active site of thermolysin.
Protein Sci., 4, 1995
|
|
3FI5
 
 | | Crystal Structure of T4 Lysozyme Mutant R96W | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, Lysozyme, ... | | Authors: | Mooers, B.H.M, Matthews, B.W. | | Deposit date: | 2008-12-11 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Contributions of all 20 amino acids at site 96 to the stability and structure of T4 lysozyme.
Protein Sci., 18, 2009
|
|
1LNC
 
 | | A STRUCTURAL ANALYSIS OF METAL SUBSTITUTIONS IN THERMOLYSIN | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, LYSINE, ... | | Authors: | Holland, D.R, Hausrath, A.C, Juers, D, Matthews, B.W. | | Deposit date: | 1994-05-13 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of zinc substitutions in the active site of thermolysin.
Protein Sci., 4, 1995
|
|
3F9L
 
 | |
1LNA
 
 | | A STRUCTURAL ANALYSIS OF METAL SUBSTITUTIONS IN THERMOLYSIN | | Descriptor: | CALCIUM ION, COBALT (II) ION, DIMETHYL SULFOXIDE, ... | | Authors: | Holland, D.R, Hausrath, A.C, Juers, D, Matthews, B.W. | | Deposit date: | 1994-05-13 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of zinc substitutions in the active site of thermolysin.
Protein Sci., 4, 1995
|
|
3GCT
 
 | |
1L02
 
 | |
1L09
 
 | |
1L06
 
 | |
1L10
 
 | |
1L15
 
 | |
2OE9
 
 | | High-pressure structure of pseudo-WT T4 Lysozyme | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2006-12-28 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Rigidity of a Large Cavity-containing Protein Revealed by High-pressure Crystallography.
J.Mol.Biol., 367, 2007
|
|
1L13
 
 | |
