8IM0
 
 | | mCherry-LaM8 complex | | Descriptor: | LaM8, MCherry fluorescent protein | | Authors: | Liang, H, Liu, R, Ding, Y. | | Deposit date: | 2023-03-05 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural Insights into the Binding of Red Fluorescent Protein mCherry-Specific Nanobodies.
Int J Mol Sci, 24, 2023
|
|
8ILX
 
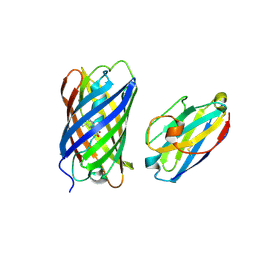 | | mCherry-LaM3 complex | | Descriptor: | LAM3, MCherry fluorescent protein | | Authors: | Liang, H, Liu, R, Ding, Y. | | Deposit date: | 2023-03-05 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural Insights into the Binding of Red Fluorescent Protein mCherry-Specific Nanobodies.
Int J Mol Sci, 24, 2023
|
|
8IM1
 
 | | mCherry-LaM1 complex | | Descriptor: | LaM1, MCherry fluorescent protein, SULFATE ION | | Authors: | Liang, H, Liu, R, Ding, Y. | | Deposit date: | 2023-03-05 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insights into the Binding of Red Fluorescent Protein mCherry-Specific Nanobodies.
Int J Mol Sci, 24, 2023
|
|
9DAK
 
 | |
4GA7
 
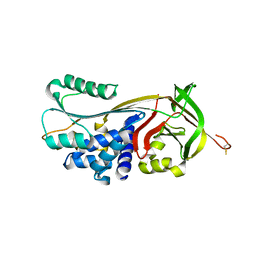 | | Crystal structure of human serpinB1 mutant | | Descriptor: | Leukocyte elastase inhibitor | | Authors: | Wang, L, Li, Q, Wu, L, Zhang, K, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of SERPINB1 as a physiological inhibitor of human granzyme H
J.Immunol., 190, 2013
|
|
4GAW
 
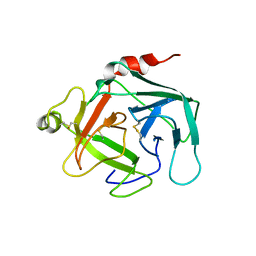 | | Crystal structure of active human granzyme H | | Descriptor: | CHLORIDE ION, Granzyme H, SULFATE ION | | Authors: | Wang, L, Li, Q, Wu, L, Zhang, K, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of SERPINB1 as a physiological inhibitor of human granzyme H
J.Immunol., 190, 2013
|
|
7ESD
 
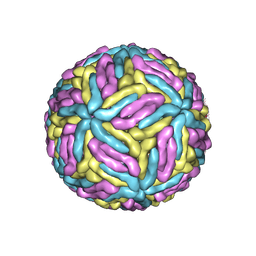 | | Mature Donggang virus | | Descriptor: | Genome polyprotein | | Authors: | Zhang, Y, Liang, D. | | Deposit date: | 2021-05-10 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Replication is the key barrier during the dual-host adaptation of mosquito-borne flaviviruses.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DGG
 
 | | Structure of glycosylated LAG-3 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Silberstein, J.L, Mathews, I.I, Frank, J.A, Chan, K.-W, Fernandez, D, Du, J, Wang, J, Kong, X.-P, Cochran, J.R. | | Deposit date: | 2022-06-23 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.78 Å) | | Cite: | Structural insights reveal interplay between LAG-3 homodimerization, ligand binding, and function.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5IFG
 
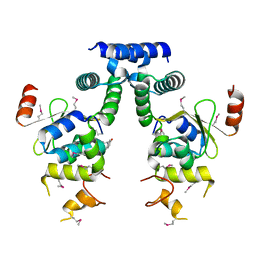 | | Crystal structure of HigA-HigB complex from E. Coli | | Descriptor: | Antitoxin HigA, mRNA interferase HigB | | Authors: | Yang, J.S, Zhou, K, Gao, z.Q, Liu, Q.S, Dong, Y.H. | | Deposit date: | 2016-02-26 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structural insight into the E. coli HigBA complex
Biochem. Biophys. Res. Commun., 478, 2016
|
|
6M1V
 
 | |
8H83
 
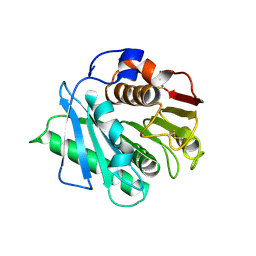 | | Crystal structure of a IsPETase variant V22 from Ideonella sakaiensis | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Wei, H.L, Gao, S.F, Li, Q, Han, X, Gao, J, Liu, W.D. | | Deposit date: | 2022-10-21 | | Release date: | 2024-04-24 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | beta-sheet Engineering of IsPETase for PET Depolymerization
Engineering (Beijing), 2024
|
|
2YU2
 
 | | Crystal structure of hJHDM1A without a-ketoglutarate | | Descriptor: | FE (II) ION, JmjC domain-containing histone demethylation protein 1A | | Authors: | Han, Z. | | Deposit date: | 2007-04-05 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for histone demethylation by JHDM1
To be Published
|
|
2YU1
 
 | | Crystal structure of hJHDM1A complexed with a-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1A | | Authors: | Han, Z. | | Deposit date: | 2007-04-05 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for histone demethylation by JHDM1
To be Published
|
|
8H92
 
 | |
6L8Z
 
 | | Crystal structure of ugt transferase mutant in complex with UPG | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2019-11-07 | | Release date: | 2020-04-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Efficient O-Glycosylation of Triterpenes Enabled by Protein Engineering of Plant Glycosyltransferase UGT74AC1
Acs Catalysis, 2020
|
|
8J5N
 
 | | Crystal structure of a PETase variant V20 from Ideonella sakaiensis | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Wei, H.L, Gao, S.F, Li, Q, Liu, W.D, Zhu, L.L. | | Deposit date: | 2023-04-23 | | Release date: | 2024-04-24 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | beta-sheet Engineering of IsPETase for PET Depolymerization
Engineering (Beijing), 2024
|
|
7WED
 
 | |
7WEA
 
 | |
7WE8
 
 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv265 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 265, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
7WE7
 
 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv282 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 282, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
7WEE
 
 | |
7WEB
 
 | |
7WEF
 
 | |
7WEC
 
 | |
7WE9
 
 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv289 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
