6DYK
 
 | |
6DY8
 
 | |
6DYI
 
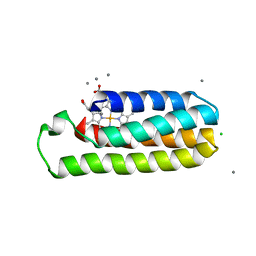 | | Co(II)-bound structure of the engineered cyt cb562 variant, H3 | | Descriptor: | CALCIUM ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Tezcan, F.A, Rittle, J. | | Deposit date: | 2018-07-01 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.964 Å) | | Cite: | An efficient, step-economical strategy for the design of functional metalloproteins.
Nat.Chem., 11, 2019
|
|
6DYB
 
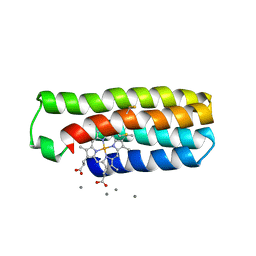 | |
6DYJ
 
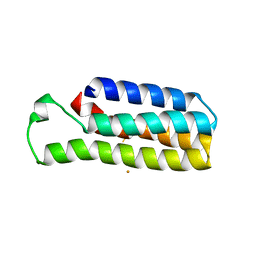 | |
6DYD
 
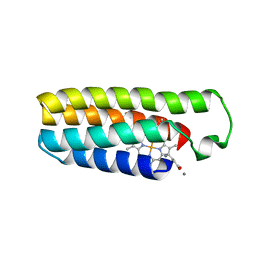 | | Cu(II)-bound structure of the engineered cyt cb562 variant, CH3 | | Descriptor: | CALCIUM ION, COPPER (II) ION, HEME C, ... | | Authors: | Tezcan, F.A, Rittle, J. | | Deposit date: | 2018-07-01 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.724 Å) | | Cite: | An efficient, step-economical strategy for the design of functional metalloproteins.
Nat.Chem., 11, 2019
|
|
6DYC
 
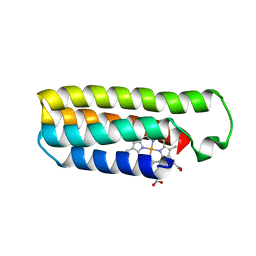 | | Co(II)-bound structure of the engineered cyt cb562 variant, CH3 | | Descriptor: | CALCIUM ION, COBALT (II) ION, HEME C, ... | | Authors: | Tezcan, F.A, Rittle, J. | | Deposit date: | 2018-07-01 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | An efficient, step-economical strategy for the design of functional metalloproteins.
Nat.Chem., 11, 2019
|
|
6DY6
 
 | | Mn(II)-bound structure of the engineered cyt cb562 variant, CH2E | | Descriptor: | HEME C, MANGANESE (II) ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Tezcan, F.A, Rittle, J. | | Deposit date: | 2018-07-01 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An efficient, step-economical strategy for the design of functional metalloproteins.
Nat.Chem., 11, 2019
|
|
6DYE
 
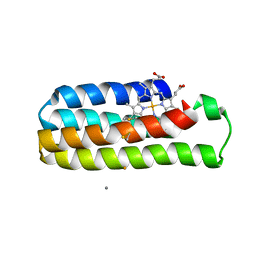 | | Fe(II)-bound structure of the engineered cyt cb562 variant, CH3 | | Descriptor: | CALCIUM ION, FE (III) ION, HEME C, ... | | Authors: | Tezcan, F.A, Rittle, J. | | Deposit date: | 2018-07-01 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | An efficient, step-economical strategy for the design of functional metalloproteins.
Nat.Chem., 11, 2019
|
|
6DYL
 
 | |
6DYG
 
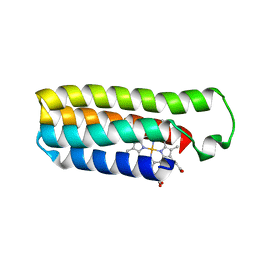 | | Fe(II)-bound structure of the engineered cyt cb562 variant, CH3Y | | Descriptor: | FE (III) ION, HEME C, MAGNESIUM ION, ... | | Authors: | Tezcan, F.A, Rittle, J. | | Deposit date: | 2018-07-01 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | An efficient, step-economical strategy for the design of functional metalloproteins.
Nat.Chem., 11, 2019
|
|
6DY4
 
 | |
8E3U
 
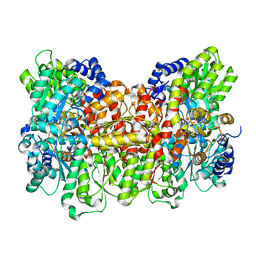 | |
8E3T
 
 | |
8E3V
 
 | |
3H3X
 
 | | Structure of the V74M large subunit mutant of NI-FE hydrogenase in an oxidized state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martinez, N, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2009-04-17 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Introduction of methionines in the gas channel makes [NiFe] hydrogenase aero-tolerant
J.Am.Chem.Soc., 131, 2009
|
|
4TNM
 
 | |
1LDG
 
 | | PLASMODIUM FALCIPARUM L-LACTATE DEHYDROGENASE COMPLEXED WITH NADH AND OXAMATE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-LACTATE DEHYDROGENASE, OXAMIC ACID | | Authors: | Dunn, C, Banfield, M, Barker, J, Higham, C, Moreton, K, Turgut-Balik, D, Brady, L, Holbrook, J.J. | | Deposit date: | 1996-09-10 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The structure of lactate dehydrogenase from Plasmodium falciparum reveals a new target for anti-malarial design.
Nat.Struct.Biol., 3, 1996
|
|
4F8C
 
 | |
1E3W
 
 | | Rat brain 3-hydroxyacyl-CoA dehydrogenase binary complex with NADH and 3-keto butyrate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETOACETIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Powell, A.J, Read, J.A, Brady, R.L. | | Deposit date: | 2000-06-26 | | Release date: | 2001-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of Structurally Diverse Substrates by Type II 3-Hydroxyacyl-Coa Dehydrogenase (Hadh II) Amyloid-Beta Binding Alcohol Dehydrogenase (Abad)
J.Mol.Biol., 303, 2000
|
|
2GL0
 
 | | Structure of PAE2307 in complex with adenosine | | Descriptor: | ADENOSINE, PHOSPHATE ION, conserved hypothetical protein | | Authors: | Lott, J.S, Paget, B, Johnston, J.M, Baker, E.N. | | Deposit date: | 2006-04-04 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure of an Ancient Conserved Domain Establishes a Structural Basis for Stable Histidine Phosphorylation and Identifies a New Family of Adenosine-specific Kinases.
J.Biol.Chem., 281, 2006
|
|
1E6W
 
 | |
1JVB
 
 | | ALCOHOL DEHYDROGENASE FROM THE ARCHAEON SULFOLOBUS SOLFATARICUS | | Descriptor: | NAD(H)-DEPENDENT ALCOHOL DEHYDROGENASE, ZINC ION | | Authors: | Esposito, L, Sica, F, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-08-29 | | Release date: | 2002-08-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the alcohol dehydrogenase from the hyperthermophilic archaeon Sulfolobus solfataricus at 1.85 A resolution.
J.Mol.Biol., 318, 2002
|
|
7RI3
 
 | | Crystal structure of Albireti Toxin, a diphtheria toxin homolog, from Streptomyces albireticuli | | Descriptor: | ACETATE ION, CALCIUM ION, Diphtheria_T domain-containing protein, ... | | Authors: | Sugiman-Marangos, S.N, Gill, S.K, Melnyk, R.A. | | Deposit date: | 2021-07-19 | | Release date: | 2022-04-20 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structures of distant diphtheria toxin homologs reveal functional determinants of an evolutionarily conserved toxin scaffold.
Commun Biol, 5, 2022
|
|
7RB4
 
 | | Crystal structure of Peptono Toxin, a diphtheria toxin homolog, from Seinonella peptonophila | | Descriptor: | Diphtheria toxin, C domain, IODIDE ION | | Authors: | Sugiman-Marangos, S.N, Gill, S.K, Melnyk, R.A. | | Deposit date: | 2021-07-05 | | Release date: | 2022-04-20 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structures of distant diphtheria toxin homologs reveal functional determinants of an evolutionarily conserved toxin scaffold.
Commun Biol, 5, 2022
|
|
