6E76
 
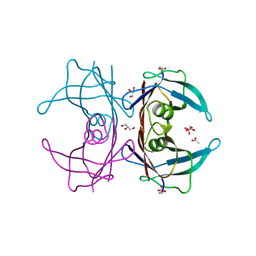 | | Structure of Human Transthyretin Asp38Ala/Thr119Met Mutant | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Saelices, L, Chung, K, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Variants of Transthyretin
To Be Published
|
|
6E75
 
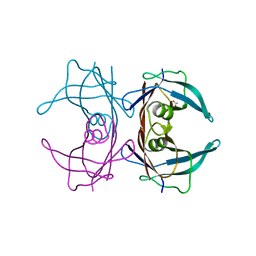 | | Structure of Human Transthyretin Asp38Ala Mutant | | Descriptor: | ACETATE ION, Transthyretin | | Authors: | Chung, K, Saelices, L, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Variants of Transthyretin
To Be Published
|
|
2ON9
 
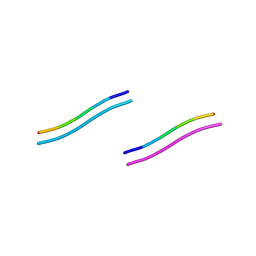 | |
2ONX
 
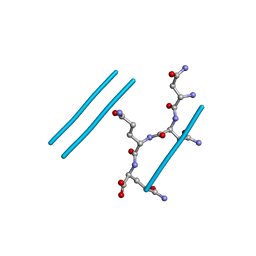 | | NNQQ peptide corresponding to residues 8-11 of yeast prion sup35 (alternate crystal form) | | Descriptor: | peptide corresponding to residues 8-11 of yeast prion sup35 | | Authors: | Sawaya, M.R, Sambashivan, S, Nelson, R, Ivanova, M, Sievers, S.A, Apostol, M.I, Thompson, M.J, Balbirnie, M, Wiltzius, J.J, McFarlane, H, Madsen, A.O, Riekel, C, Eisenberg, D. | | Deposit date: | 2007-01-24 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Atomic structures of amyloid cross-beta spines reveal varied steric zippers.
Nature, 447, 2007
|
|
6E6Z
 
 | | Structure of Wild Type Human Transthyretin in Complex with Tafamidis | | Descriptor: | 2-(3,5-dichlorophenyl)-1,3-benzoxazole-6-carboxylic acid, Transthyretin | | Authors: | Chung, K, Saelices, L, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Variants of Transthyretin
To Be Published
|
|
6E77
 
 | | Structure of Human Transthyretin Asp38Ala Mutant in Complex with Tafamidis | | Descriptor: | 2-(3,5-dichlorophenyl)-1,3-benzoxazole-6-carboxylic acid, Transthyretin | | Authors: | Saelices, L, Chung, K, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Variants of Transthyretin
To Be Published
|
|
2OKZ
 
 | |
3N3E
 
 | | Zebrafish AlphaA crystallin | | Descriptor: | Alpha A crystallin | | Authors: | Laganowsky, A, Eisenberg, D. | | Deposit date: | 2010-05-19 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Non-3D domain swapped crystal structure of truncated zebrafish alphaA crystallin.
Protein Sci., 19, 2010
|
|
2OMM
 
 | |
2OMQ
 
 | |
1F52
 
 | | CRYSTAL STRUCTURE OF GLUTAMINE SYNTHETASE FROM SALMONELLA TYPHIMURIUM CO-CRYSTALLIZED WITH ADP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLUTAMINE SYNTHETASE, ... | | Authors: | Gill, H.S, Pfluegl, G.M.U, Eisenberg, D. | | Deposit date: | 2000-06-12 | | Release date: | 2000-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The crystal structure of phosphinothricin in the active site of glutamine synthetase illuminates the mechanism of enzymatic inhibition.
Biochemistry, 40, 2001
|
|
1HTO
 
 | |
1HTQ
 
 | | Multicopy crystallographic structure of a relaxed glutamine synthetase from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE MONOPHOSPHATE, CITRIC ACID, MANGANESE (II) ION, ... | | Authors: | Gill, H.S, Pfluegl, G.M, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2001-01-01 | | Release date: | 2002-07-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Multicopy crystallographic refinement of a relaxed glutamine synthetase from Mycobacterium tuberculosis highlights flexible loops in the enzymatic mechanism and its regulation.
Biochemistry, 41, 2002
|
|
1FPY
 
 | |
1IK6
 
 | |
1G6U
 
 | | CRYSTAL STRUCTURE OF A DOMAIN SWAPPED DIMER | | Descriptor: | DOMAIN SWAPPED DIMER, SULFATE ION, trifluoroacetic acid | | Authors: | Ogihara, N.L, Ghirlanda, G, Bryson, J.W, Gingery, M, DeGrado, W.F, Eisenberg, D. | | Deposit date: | 2000-11-07 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Design of three-dimensional domain-swapped dimers and fibrous oligomers.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JBM
 
 | |
1JRI
 
 | |
1JER
 
 | | CUCUMBER STELLACYANIN, CU2+, PH 7.0 | | Descriptor: | COPPER (II) ION, CUCUMBER STELLACYANIN | | Authors: | Hart, P.J, Nersissian, A.M, Herrmann, R.G, Nalbandyan, R.M, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1996-08-21 | | Release date: | 1997-02-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A missing link in cupredoxins: crystal structure of cucumber stellacyanin at 1.6 A resolution.
Protein Sci., 5, 1996
|
|
3G7V
 
 | | Islet Amyloid Polypeptide (IAPP or Amylin) fused to Maltose Binding Protein | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein, Islet amyloid polypeptide fusion protein, ... | | Authors: | Wiltzius, J.J.W, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2009-02-10 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Atomic structures of IAPP (amylin) fusions suggest a mechanism for fibrillation and the role of insulin in the process
Protein Sci., 18, 2009
|
|
3HYD
 
 | |
4PM4
 
 | | Structure of a putative periplasmic iron siderophore binding protein (Rv0265c) from Mycobacterium tuberculosis H37Rv | | Descriptor: | CHLORIDE ION, Iron complex transporter substrate-binding protein, SULFATE ION | | Authors: | Arbing, M.A, Chan, S, Tran, N, Kuo, E, Lu, J, Harris, L.R, Zhou, T.T, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-05-20 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a putative periplasmic iron siderophore binding protein (Rv0265c) from Mycobacterium tuberculosis H37Rv
To Be Published
|
|
3G7W
 
 | | Islet Amyloid Polypeptide (IAPP or Amylin) Residues 1 to 22 fused to Maltose Binding Protein | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein, Islet amyloid polypeptide fusion protein, ... | | Authors: | Wiltzius, J.J.W, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2009-02-11 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Atomic structures of IAPP (amylin) fusions suggest a mechanism for fibrillation and the role of insulin in the process
Protein Sci., 18, 2009
|
|
1LMI
 
 | | 1.5 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF A SECRETED PROTEIN FROM MYCOBACTERIUM TUBERCULOSIS-MPT63 | | Descriptor: | Immunogenic protein MPT63/MPB63 | | Authors: | Goulding, C.W, Parseghian, A, Sawaya, M.R, Cascio, D, Apostol, M, Gennaro, M.L, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-05-01 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a major secreted protein of Mycobacterium tuberculosis-MPT63 at
1.5-A resolution
Protein Sci., 11, 2002
|
|
3LOW
 
 | |
