2RVK
 
 | |
6LXV
 
 | | Cryo-EM structure of phosphoketolase from Bifidobacterium longum | | Descriptor: | CALCIUM ION, Phosphoketolase, THIAMINE DIPHOSPHATE | | Authors: | Nakata, K, Miyazaki, N, Yamaguchi, H, Hirose, M, Miyano, H, Mizukoshi, T, Kashiwagi, T, Iwasaki, K. | | Deposit date: | 2020-02-12 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | High-resolution structure of phosphoketolase from Bifidobacterium longum determined by cryo-EM single-particle analysis.
J.Struct.Biol., 214, 2022
|
|
4E9X
 
 | | Multicopper Oxidase mgLAC (data3) | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Multicopper oxidase, ... | | Authors: | Komori, H, Miyazaki, K, Higuchi, Y. | | Deposit date: | 2012-03-21 | | Release date: | 2013-03-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | New insights into the catalytic active-site structure of multicopper oxidases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4E9V
 
 | | Multicopper Oxidase mgLAC (data1) | | Descriptor: | CHLORIDE ION, COPPER (II) ION, HYDROXIDE ION, ... | | Authors: | Komori, H, Miyazaki, K, Higuchi, Y. | | Deposit date: | 2012-03-21 | | Release date: | 2013-03-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New insights into the catalytic active-site structure of multicopper oxidases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4XM0
 
 | | N,N'-diacetylchitobiose deacetylase (SeMet derivative) from Pyrococcus furiosus in the absence of cadmium | | Descriptor: | Uncharacterized protein, ZINC ION | | Authors: | Nakamura, T, Niiyama, M, Hashimoto, W, Ida, K, Uegaki, K. | | Deposit date: | 2015-01-14 | | Release date: | 2015-06-10 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Multiple crystal forms of N,N'-diacetylchitobiose deacetylase from Pyrococcus furiosus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4XM2
 
 | | N,N'-diacetylchitobiose deacetylase from Pyrococcus furiosus in the absence of cadmium | | Descriptor: | Uncharacterized protein, ZINC ION | | Authors: | Nakamura, T, Niiyama, M, Hashimoto, W, Ida, K, Uegaki, K. | | Deposit date: | 2015-01-14 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple crystal forms of N,N'-diacetylchitobiose deacetylase from Pyrococcus furiosus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4XLZ
 
 | | N,N'-diacetylchitobiose deacetylase (SeMet derivative) from Pyrococcus furiosus in the presence of cadmium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Nakamura, T, Niiyama, M, Hashimoto, W, Ida, K, Uegaki, K. | | Deposit date: | 2015-01-14 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Multiple crystal forms of N,N'-diacetylchitobiose deacetylase from Pyrococcus furiosus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4XM1
 
 | | N,N'-diacetylchitobiose deacetylase from Pyrococcus furiosus in the presence of cadmium | | Descriptor: | CADMIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nakamura, T, Niiyama, M, Hashimoto, W, Ida, K, Uegaki, K. | | Deposit date: | 2015-01-14 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multiple crystal forms of N,N'-diacetylchitobiose deacetylase from Pyrococcus furiosus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
1IRG
 
 | | INTERFERON REGULATORY FACTOR-2 DNA BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | INTERFERON REGULATORY FACTOR-2 | | Authors: | Furui, J, Uegaki, K, Yamazaki, T, Shirakawa, M, Swindells, M.B, Harada, H, Taniguchi, T, Kyogoku, Y. | | Deposit date: | 1997-11-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the IRF-2 DNA-binding domain: a novel subgroup of the winged helix-turn-helix family.
Structure, 6, 1998
|
|
5ZFS
 
 | | Crystal structure of Arthrobacter globiformis M30 sugar epimerase which can produce D-allulose from D-fructose | | Descriptor: | ACETATE ION, D-allulose-3-epimerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yoshihara, A, Gullapalli, P.K, Ohtani, K, Akimitsu, K, Izumori, K, Kamitori, S. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray structure of Arthrobacter globiformis M30 ketose 3-epimerase for the production of D-allulose from D-fructose.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
2SXL
 
 | | SEX-LETHAL RBD1, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | SEX-LETHAL PROTEIN | | Authors: | Inoue, M, Muto, Y, Sakamoto, H, Kigawa, T, Takio, K, Shimura, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1997-07-16 | | Release date: | 1998-07-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A characteristic arrangement of aromatic amino acid residues in the solution structure of the amino-terminal RNA-binding domain of Drosophila sex-lethal.
J.Mol.Biol., 272, 1997
|
|
5B2E
 
 | | N,N'-diacetylchitobiose deacetylase from Pyrococcus horikoshii complexed with its inhibitor MPG (acetate-containing condition) | | Descriptor: | 2-deoxy-2-{[(S)-hydroxy(methyl)phosphoryl]amino}-beta-D-glucopyranose, HEXANE-1,6-DIOL, Putative uncharacterized protein PH0499, ... | | Authors: | Nakamura, T, Niiyama, M, Ida, K, Uegaki, K. | | Deposit date: | 2016-01-15 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate recognition of N,N'-diacetylchitobiose deacetylase from Pyrococcus horikoshii
J.Struct.Biol., 195, 2016
|
|
5AXV
 
 | | Crystal Structure of Cypovirus Polyhedra R13K Mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Abe, S, Ijiri, H, Negishi, H, Yamanaka, H, Sasaki, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2015-08-01 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Design of Enzyme-Encapsulated Protein Containers by In Vivo Crystal Engineering
Adv. Mater. Weinheim, 27, 2015
|
|
1UKJ
 
 | | Detailed structure of L-Methionine-Lyase from Pseudomonas putida | | Descriptor: | Methionine gamma-lyase, SULFATE ION | | Authors: | Misaki, S, Takimoto, A, Takakura, T, Yoshioka, T, Yamashita, M, Tamura, T, Tanaka, H, Inagaki, K. | | Deposit date: | 2003-08-24 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Detailed structure of L-Methionine -Lyase from Pseudomonas putida
To be Published
|
|
5B2F
 
 | | N,N'-diacetylchitobiose deacetylase from Pyrococcus horikoshii complexed with its inhibitor MPG (phosphate-containing condition) | | Descriptor: | 2-deoxy-2-{[(S)-hydroxy(methyl)phosphoryl]amino}-beta-D-glucopyranose, Putative uncharacterized protein PH0499, ZINC ION | | Authors: | Nakamura, T, Niiyama, M, Ida, K, Uegaki, K. | | Deposit date: | 2016-01-15 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate recognition of N,N'-diacetylchitobiose deacetylase from Pyrococcus horikoshii
J.Struct.Biol., 195, 2016
|
|
1IRF
 
 | | INTERFERON REGULATORY FACTOR-2 DNA BINDING DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | INTERFERON REGULATORY FACTOR-2 | | Authors: | Furui, J, Uegaki, K, Yamazaki, T, Shirakawa, M, Swindells, M.B, Harada, H, Taniguchi, T, Kyogoku, Y. | | Deposit date: | 1997-11-24 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the IRF-2 DNA-binding domain: a novel subgroup of the winged helix-turn-helix family.
Structure, 6, 1998
|
|
6A7P
 
 | | Human serum albumin complexed with aripiprazole | | Descriptor: | 7-[4-[4-[2,3-bis(chloranyl)phenyl]piperazin-1-yl]butoxy]-3,4-dihydro-1H-quinolin-2-one, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Kawai, A, Yamasaki, K, Otagiri, M. | | Deposit date: | 2018-07-03 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Analysis of the Binding of Aripiprazole to Human Serum Albumin: The Importance of a Chloro-Group in the Chemical Structure.
Acs Omega, 3, 2018
|
|
7Y51
 
 | | Acetylxylan esterase from Caldanaerobacter subterraneus subsp. tengcongensis TTE0866 delta100 mutant | | Descriptor: | GLYCEROL, NICKEL (II) ION, Predicted xylanase/chitin deacetylase | | Authors: | Sasamoto, K, Himiyama, T, Moriyoshi, K, Ohmoto, T, Uegaki, K, Nakamura, T, Nishiya, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional analysis of the N-terminal region of acetylxylan esterase from Caldanaerobacter subterraneus subsp. tengcongensis.
Febs Open Bio, 12, 2022
|
|
3W80
 
 | |
3W7Y
 
 | |
5B59
 
 | | Hen egg-white lysozyme modified with a keto-ABNO. | | Descriptor: | (2~{S})-2-azanyl-3-[(2~{R},3~{S})-2-oxidanyl-3-[[(1~{S},5~{R})-3-oxidanylidene-9-azabicyclo[3.3.1]nonan-9-yl]oxy]-1,2-dihydroindol-3-yl]propanal, Lysozyme C | | Authors: | Sasaki, D, Seki, Y, Sohma, Y, Oisaki, K, Kanai, M. | | Deposit date: | 2016-04-28 | | Release date: | 2016-09-14 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Transition Metal-Free Tryptophan-Selective Bioconjugation of Proteins
J.Am.Chem.Soc., 138, 2016
|
|
2LMK
 
 | | Solution Structure of Mouse Pheromone ESP1 | | Descriptor: | Exocrine gland-secreting peptide 1 | | Authors: | Yoshinaga, S, Sato, T, Hirakane, M, Esaki, K, Hamaguchi, T, Haga-Yamanaka, S, Tsunoda, M, Kimoto, H, Shimada, I, Touhara, K, Terasawa, H. | | Deposit date: | 2011-12-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the Mouse Sex Peptide Pheromone ESP1 Reveals a Molecular Basis for Specific Binding to the Class-C G-Protein-Coupled Vomeronasal Receptor
J.Biol.Chem., 2013
|
|
3W7Z
 
 | |
6IU6
 
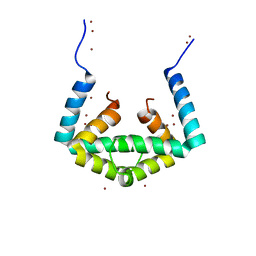 | | Crystal structure of cytoplasmic metal binding domain with nickel ions | | Descriptor: | NICKEL (II) ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
6IU3
 
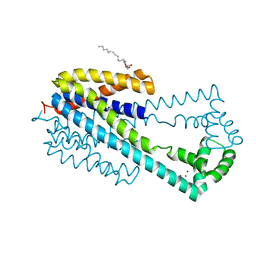 | | Crystal structure of iron transporter VIT1 with zinc ions | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Taniguchi, R, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
