7AR9
 
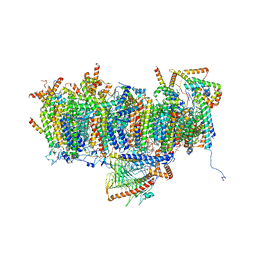 | | Cryo-EM structure of Polytomella Complex-I (membrane arm) | | Descriptor: | (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 15 kDa, AGGG, ... | | Authors: | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7ARD
 
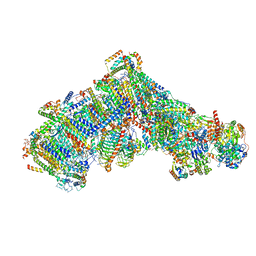 | | Cryo-EM structure of Polytomella Complex-I (complete composition) | | Descriptor: | (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 13 kDa, 15 kDa, ... | | Authors: | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
5AOE
 
 | |
5AOF
 
 | |
4B2Q
 
 | |
7O71
 
 | | Cryo-EM structure of a respiratory complex I | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Acyl carrier protein ACPM1 of NADH:Ubiquinone Oxidoreductase (Complex I), ... | | Authors: | Parey, K, Vonck, J. | | Deposit date: | 2021-04-12 | | Release date: | 2021-11-10 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | High-resolution structure and dynamics of mitochondrial complex I-Insights into the proton pumping mechanism.
Sci Adv, 7, 2021
|
|
7O6Y
 
 | | Cryo-EM structure of respiratory complex I under turnover | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Parey, K, Vonck, J. | | Deposit date: | 2021-04-12 | | Release date: | 2021-11-10 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | High-resolution structure and dynamics of mitochondrial complex I-Insights into the proton pumping mechanism.
Sci Adv, 7, 2021
|
|
6RFR
 
 | | Cryo-EM structure of respiratory complex I from Yarrowia lipolytica at 3.2 A resolution | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Acyl carrier protein ACPM1 of NADH:Ubiquinone Oxidoreductase (Complex I), ... | | Authors: | Parey, K, Vonck, J. | | Deposit date: | 2019-04-16 | | Release date: | 2019-12-11 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution cryo-EM structures of respiratory complex I: Mechanism, assembly, and disease.
Sci Adv, 5, 2019
|
|
6RFS
 
 | | Cryo-EM structure of a respiratory complex I mutant lacking NDUFS4 | | Descriptor: | Acyl carrier protein ACPM1 of NADH:Ubiquinone Oxidoreductase (Complex I), Acyl carrier protein ACPM2 of NADH:Ubiquinone Oxidoreductase (Complex I), FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Parey, K, Vonck, J. | | Deposit date: | 2019-04-16 | | Release date: | 2019-12-11 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | High-resolution cryo-EM structures of respiratory complex I: Mechanism, assembly, and disease.
Sci Adv, 5, 2019
|
|
6TA1
 
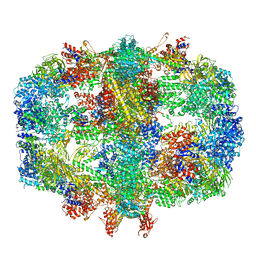 | | Fatty acid synthase of S. cerevisiae | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta, ... | | Authors: | Vonck, J, D'Imprima, E, Joppe, M, Grininger, M. | | Deposit date: | 2019-10-29 | | Release date: | 2019-11-06 | | Last modified: | 2020-05-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The resolution revolution in cryoEM requires high-quality sample preparation: a rapid pipeline to a high-resolution map of yeast fatty acid synthase.
Iucrj, 7, 2020
|
|
7ZMB
 
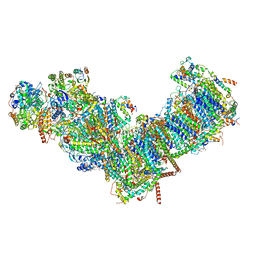 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 2) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
7ZMG
 
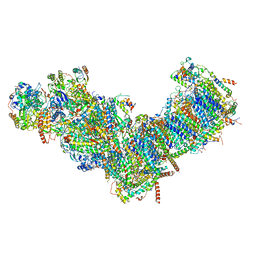 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 1) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
7ZM7
 
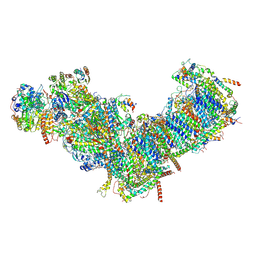 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (inhibited by DDM) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
7ZME
 
 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 2) - membrane arm | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
7ZMH
 
 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 1) - membrane arm | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
7ZM8
 
 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (inhibited by DDM) - membrane arm | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
8B4I
 
 | | Cryo-EM structure of the Neurospora crassa TOM core complex at 3.3 angstrom | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, DIUNDECYL PHOSPHATIDYL CHOLINE, Mitochondrial import receptor subunit Tom22, ... | | Authors: | Ornelas, P, Kuehlbrandt, W. | | Deposit date: | 2022-09-20 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Two conformations of the Tom20 preprotein receptor in the TOM holo complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8BEL
 
 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CIII membrane domain) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-11 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8BEP
 
 | |
8BEF
 
 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CI membrane core) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-11 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8BEE
 
 | |
8BED
 
 | |
8BEH
 
 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CI membrane tip) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-11 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8BPX
 
 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Complete composition) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-11-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.09 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8BQ5
 
 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Complete conformation 1 composition) | | Descriptor: | 2,3-DIMETHOXY-5-METHYL-6-(3,11,15,19-TETRAMETHYL-EICOSA-2,6,10,14,18-PENTAENYL)-[1,4]BENZOQUINONE, AT3G07480.1, Acyl carrier protein 1, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-11-18 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
