8I8P
 
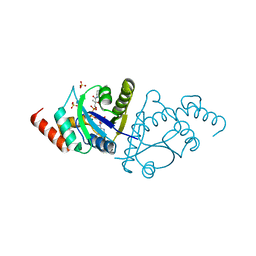 | | Crystal structure of the complex of phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Dephosphocoenzyme-A at 2.19 A resolution. | | Descriptor: | CHLORIDE ION, DEPHOSPHO COENZYME A, MAGNESIUM ION, ... | | Authors: | Ahmad, N, Viswanathan, V, Gupta, A, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2023-02-04 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of the complex of phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Dephosphocoenzyme-A at 2.19 A resolution.
To Be Published
|
|
7WGK
 
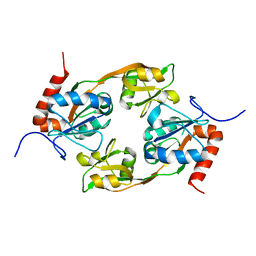 | | Crystal structure of Adenosine triphosphate phosphoribosyltransferase (HisG) from Acinetobacter baumannii at 3.13 A resolution | | Descriptor: | ATP phosphoribosyltransferase | | Authors: | Ahmad, N, Maurya, A, Singh, P.K, Viswanathan, V, Sharma, P, Singh, T.P, Sharma, S. | | Deposit date: | 2021-12-28 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Crystal structure of Adenosine triphosphate phosphoribosyltransferase (HisG) from Acinetobacter baumannii at 3.13 A resolution
To Be Published
|
|
7WGM
 
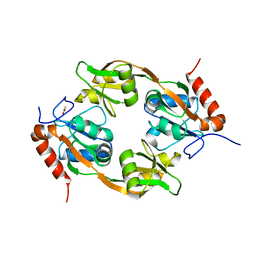 | | Crystal structure of Adenosine triphosphate phosphoribosyltransferase (HisG) from Acinetobacter baumannii at 3.15 A resolution | | Descriptor: | ACETATE ION, ATP phosphoribosyltransferase | | Authors: | Ahmad, N, Maurya, A, Singh, P.K, Viswanathan, V, Sharma, P, Singh, T.P, Sharma, S. | | Deposit date: | 2021-12-28 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of Adenosine triphosphate phosphoribosyltransferase (HisG) from Acinetobacter baumannii at 3.15 A resolution
To Be Published
|
|
3NNO
 
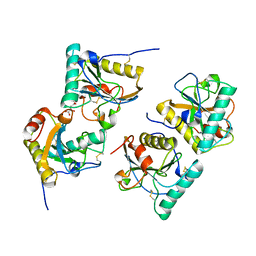 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with Alpha-Rhamnose at 2.9 A resolution | | Descriptor: | L(+)-TARTARIC ACID, Peptidoglycan recognition protein 1, alpha-L-rhamnopyranose | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-24 | | Release date: | 2010-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with Alpha-Rhamnose at 2.9 A resolution
To be Published
|
|
3QV4
 
 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with dipeptide L-ALA D-GLU at 2.7 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALANINE, ... | | Authors: | Shukla, P.K, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-02-25 | | Release date: | 2011-03-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with dipeptide L-ALA D-GLU at 2.7 A resolution
To be Published
|
|
3QS0
 
 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with a bound N-acetylglucosamine in the diffusion channel at 2.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-02-19 | | Release date: | 2011-03-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with a bound N-acetylglucosamine in the diffusion channel AT 2.5 A resolution
To be Published
|
|
9IIL
 
 | | Structure of the complex of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii with nicotinamide adenine dinucleotide in the presence of poly(ethylene glycol) at 2.20 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Jeyakanthan, J, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-06-20 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the complex of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii with nicotinamide adenine dinucleotide in the presence of poly(ethylene glycol) at 2.20 A resolution
To Be Published
|
|
9IYE
 
 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.39 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-07-30 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.39 A resolution.
To Be Published
|
|
9IYG
 
 | |
9IYF
 
 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.37 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-07-30 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.37 A resolution.
To Be Published
|
|
9IYH
 
 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.25 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-07-30 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.25 A resolution.
To Be Published
|
|
9IT8
 
 | | Crystal structure of the ternary complex of lactoperoxidase with nitric oxide and nitrite ion at 1.95 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maurya, A, Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-07-19 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Crystal structure of the ternary complex of lactoperoxidase with nitric oxide and nitrite ion at 1.95 A resolution
To Be Published
|
|
9IIM
 
 | | Structure of the complex of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii with nicotinamide adenine dinucleotide at 2.74 A resolution. | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Jeyakanthan, J, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-06-20 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of the complex of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii with nicotinamide adenine dinucleotide at 2.74 A resolution.
To Be Published
|
|
8ZN1
 
 | | Structure of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 3.00 A resolution | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 3.00 A resolution
To Be Published
|
|
8ZN4
 
 | | Crystal structure of Poly(ethylene glycol) stabilized erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 2.30 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, ... | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Poly(ethylene glycol) stabilized erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 2.30 A resolution
To Be Published
|
|
9IJ6
 
 | | Crystal structure of the complex of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii with Adenosine phosphate at 2.40 A resolution. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glyceraldehyde-3-phosphate dehydrogenase, MAGNESIUM ION, ... | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Jeyakanthan, J, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-06-21 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the complex of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii with Adenosine phosphate at 2.40 A resolution.
To Be Published
|
|
8ZN2
 
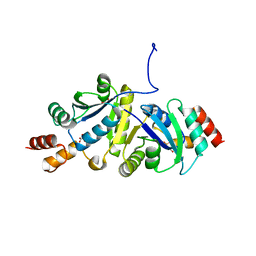 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.65 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Bhushan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.65 A resolution.
To Be Published
|
|
8ZN3
 
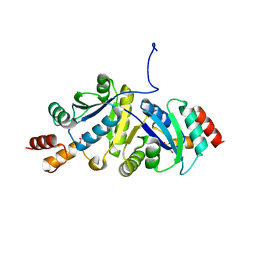 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.41 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Bhushan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.41 A resolution.
To Be Published
|
|
6LSO
 
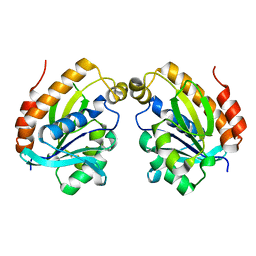 | |
7EV0
 
 | | Crystal structure of pepsin cleaved C-terminal half of lactoferrin at 2.7A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Singh, J, Ahmad, M.I, Maurya, A, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of pepsin cleaved C-terminal half of lactoferrin at 2.7A resolution
To Be Published
|
|
7EVQ
 
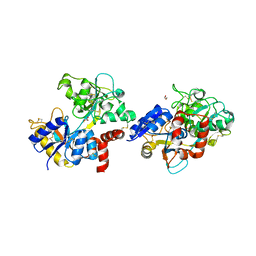 | | Crystal structure of C-terminal half of lactoferrin obtained by limited proteolysis using pepsin at 2.6 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, ... | | Authors: | Viswanathan, V, Singh, J, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2021-05-21 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of C-terminal half of lactoferrin obtained by limited proteolysis using pepsin at 2.6 A resolution
To Be Published
|
|
7FDW
 
 | | Crystal structure of pepsin cleaved lactoferrin C-lobe at 2.28 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Singh, P.K, Singh, J, Maurya, A, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2021-07-18 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.277 Å) | | Cite: | A Peptide Bond from the Inter-lobe Segment in the Bilobal Lactoferrin Acts as a Preferred Site for Cleavage for Serine Proteases to Generate the Perfect C-lobe: Structure of the Pepsin Hydrolyzed Lactoferrin C-lobe at 2.28 angstrom Resolution.
Protein J., 40, 2021
|
|
7WZY
 
 | | Crystal structure of Adenosine triphosphate phosphoribosyltransferase (HisG) from Acinetobacter baumannii at 2.975 A resolution | | Descriptor: | ATP phosphoribosyltransferase, FORMIC ACID, GLYCEROL | | Authors: | Ahmad, N, Maurya, A, Singh, P.K, Viswanathan, V, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.975 Å) | | Cite: | Crystal structure of Adenosine triphosphate phosphoribosyltransferase (HisG) from Acinetobacter baumannii at 2.975 A resolution
To Be Published
|
|
7XFX
 
 | | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.28 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Maurya, A, Singh, P.K, Viswanathan, V, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.28 A resolution.
To Be Published
|
|
7XFY
 
 | | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.67 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Maurya, A, Singh, P.K, Viswanathan, V, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.67 A resolution.
To Be Published
|
|
