8B29
 
 | | Human carbonic anhydrase II containing 6-fluorotryptophanes. | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Pham, L.B.T, Costantino, A, Barbieri, L, Calderone, V, Luchinat, E, Banci, L. | | Deposit date: | 2022-09-13 | | Release date: | 2023-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Direct Expression of Fluorinated Proteins in Human Cells for 19 F In-Cell NMR Spectroscopy.
J.Am.Chem.Soc., 145, 2023
|
|
2AXX
 
 | | THE SOLUTION STRUCTURE OF OXIDIZED RAT MICROSOMAL CYTOCHROME B5, NMR, 21 STRUCTURES | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnesano, F, Banci, L, Bertini, I, Felli, I.C. | | Deposit date: | 1997-10-22 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of oxidized rat microsomal cytochrome b5.
Biochemistry, 37, 1998
|
|
7NXH
 
 | | Structure of SARS-CoV2 NSP5 (3C-like proteinase) determined in-house | | Descriptor: | 3C-like proteinase | | Authors: | Calderone, V, Grifagni, D, Cantini, F, Fragai, M, Banci, L. | | Deposit date: | 2021-03-18 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SARS-CoV-2 M pro inhibition by a zinc ion: structural features and hints for drug design.
Chem.Commun.(Camb.), 57, 2021
|
|
7NWX
 
 | | SARS-COV2 NSP5 in the presence of Zn2+ | | Descriptor: | Replicase polyprotein 1a, ZINC ION | | Authors: | Calderone, V, Grifagni, D, Cantini, F, Fragai, M, Banci, L. | | Deposit date: | 2021-03-17 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SARS-CoV-2 M pro inhibition by a zinc ion: structural features and hints for drug design.
Chem.Commun.(Camb.), 57, 2021
|
|
8B0T
 
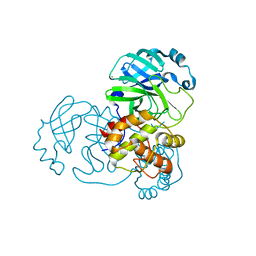 | | SARS-CoV-2 Main Protease adduct with Au(PEt3)Br | | Descriptor: | 3C-like proteinase nsp5, GOLD ION | | Authors: | Massai, L, Grifagni, D, De Santis, A, Geri, A, Calderone, V, Cantini, F, Banci, L, Messori, L. | | Deposit date: | 2022-09-08 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Gold-Based Metal Drugs as Inhibitors of Coronavirus Proteins: The Inhibition of SARS-CoV-2 Main Protease by Auranofin and Its Analogs.
Biomolecules, 12, 2022
|
|
8B0S
 
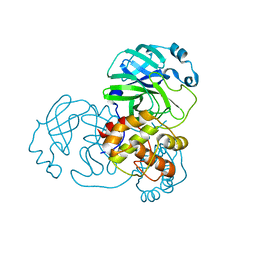 | | SARS-COV-2 Main Protease adduct with Au(NHC)Cl | | Descriptor: | 3C-like proteinase nsp5, GOLD ION | | Authors: | Massai, L, Grifagni, D, Desantis, A, Geri, A, Calderone, V, Cantini, F, Messori, L, Banci, L. | | Deposit date: | 2022-09-08 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Gold-Based Metal Drugs as Inhibitors of Coronavirus Proteins: The Inhibition of SARS-CoV-2 Main Protease by Auranofin and Its Analogs.
Biomolecules, 12, 2022
|
|
6QE4
 
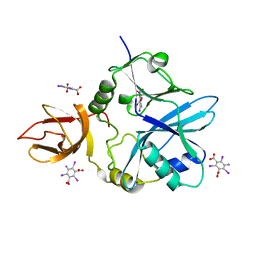 | | Re-refinement of 5OLI human IBA57-I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Putative transferase CAF17, mitochondrial | | Authors: | Calderone, V, Ciofi-Baffoni, S, Gourdoupis, S, Banci, L, Nasta, V. | | Deposit date: | 2019-01-04 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In-house high-energy-remote SAD phasing using the magic triangle: how to tackle the P1 low symmetry using multiple orientations of the same crystal of human IBA57 to increase the multiplicity.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6QE3
 
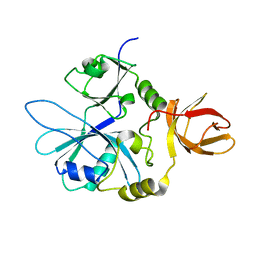 | | Re-refinement of 6ESR human IBA57 at 1.75 A resolution | | Descriptor: | Putative transferase CAF17, mitochondrial | | Authors: | Calderone, V, Ciofi-Baffoni, S, Gourdoupis, S, Banci, L. | | Deposit date: | 2019-01-04 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | In-house high-energy-remote SAD phasing using the magic triangle: how to tackle the P1 low symmetry using multiple orientations of the same crystal of human IBA57 to increase the multiplicity.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
2RML
 
 | | Solution structure of the N-terminal soluble domains of Bacillus subtilis CopA | | Descriptor: | Copper-transporting P-type ATPase copA | | Authors: | Singleton, C, Banci, L, Bertini, I, Ciofi-Baffoni, S, Tenori, L, Kihlken, M.A, Boetzel, R, Le Brun, N.E. | | Deposit date: | 2007-10-30 | | Release date: | 2008-02-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and Cu(I)-binding properties of the N-terminal soluble domains of Bacillus subtilis CopA
Biochem.J., 411, 2008
|
|
1NEW
 
 | | Cytochrome C551.5, NMR | | Descriptor: | CYTOCHROME C551.5, HEME C | | Authors: | Assfalg, M, Banci, L, Bertini, I, Bruschi, M, Turano, P. | | Deposit date: | 1998-02-10 | | Release date: | 1998-04-29 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | 800 MHz 1H NMR solution structure refinement of oxidized cytochrome c7 from Desulfuromonas acetoxidans.
Eur.J.Biochem., 256, 1998
|
|
1OSC
 
 | | Crystal structure of rat CUTA1 at 2.15 A resolution | | Descriptor: | similar to divalent cation tolerant protein CUTA | | Authors: | Arnesano, F, Banci, L, Benvenuti, M, Bertini, I, Calderone, V, Mangani, S, Viezzoli, M.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Evolutionarily Conserved Trimeric Structure of CutA1 Proteins
Suggests a Role in Signal Transduction
J.Biol.Chem., 278, 2003
|
|
1AXX
 
 | | THE SOLUTION STRUCTURE OF OXIDIZED RAT MICROSOMAL CYTOCHROME B5, NMR, 19 STRUCTURES | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnesano, F, Banci, L, Bertini, I, Felli, I.C. | | Deposit date: | 1997-10-22 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of oxidized rat microsomal cytochrome b5.
Biochemistry, 37, 1998
|
|
1BFX
 
 | | THE SOLUTION NMR STRUCTURE OF THE B FORM OF OXIDIZED RAT MICROSOMAL CYTOCHROME B5, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnesano, F, Banci, L, Bertini, I, Felli, I.C. | | Deposit date: | 1998-05-23 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the B form of oxidized rat microsomal cytochrome b5 and backbone dynamics via 15N rotating-frame NMR-relaxation measurements. Biological implications.
Eur.J.Biochem., 260, 1999
|
|
1BLV
 
 | | SOLUTION STRUCTURE OF OXIDIZED RAT MICROSOMAL CYTOCHROME B5 IN THE PRESENCE OF 2 M GUANIDINIUM CHLORIDE: MONITORING THE EARLY STEPS IN PROTEIN UNFOLDING | | Descriptor: | PROTEIN (CYTOCHROME B5), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnesano, F, Banci, L, Bertini, I, Koulougliotis, D. | | Deposit date: | 1998-07-21 | | Release date: | 1998-07-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of oxidized rat microsomal cytochrome b5 in the presence of 2 M guanidinium chloride: monitoring the early steps in protein unfolding.
Biochemistry, 37, 1998
|
|
1AW3
 
 | | THE SOLUTION NMR STRUCTURE OF OXIDIZED RAT MICROSOMAL CYTOCHROME B5, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnesano, F, Banci, L, Bertini, I, Felli, I.C. | | Deposit date: | 1997-10-09 | | Release date: | 1998-02-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of oxidized rat microsomal cytochrome b5.
Biochemistry, 37, 1998
|
|
1FD8
 
 | | SOLUTION STRUCTURE OF THE CU(I) FORM OF THE YEAST METALLOCHAPERONE, ATX1 | | Descriptor: | ATX1 COPPER CHAPERONE, COPPER (I) ION | | Authors: | Arnesano, F, Banci, L, Bertini, I, Huffman, D.L, O'Halloran, T.V. | | Deposit date: | 2000-07-20 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Cu(I) and apo forms of the yeast metallochaperone, Atx1.
Biochemistry, 40, 2001
|
|
1FES
 
 | | SOLUTION STRUCTURE OF THE APO FORM OF THE YEAST METALLOCHAPERONE, ATX1 | | Descriptor: | ATX1 COPPER CHAPERONE | | Authors: | Arnesano, F, Banci, L, Bertini, I, Huffman, D.L, O'Halloran, T.V. | | Deposit date: | 2000-07-22 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Cu(I) and apo forms of the yeast metallochaperone, Atx1.
Biochemistry, 40, 2001
|
|
1QQ3
 
 | | THE SOLUTION STRUCTURE OF THE HEME BINDING VARIANT ARG98CYS OF OXIDIZED ESCHERICHIA COLI CYTOCHROME B562 | | Descriptor: | CYTOCHROME B562, HEME B/C | | Authors: | Arnesano, F, Banci, L, Bertini, I, Ciofi-Baffoni, S, Barker, P.D, Woodyear, T. | | Deposit date: | 1999-06-10 | | Release date: | 2000-05-24 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structural consequences of b- to c-type heme conversion in oxidized Escherichia coli cytochrome b562.
Biochemistry, 39, 2000
|
|
2L4O
 
 | | Solution structure of the Streptococcus pneumoniae RrgB pilus backbone D1 domain | | Descriptor: | Cell wall surface anchor family protein | | Authors: | Gentile, M.A, Melchiorre, S, Emolo, C, Moschioni, M, Gianfaldoni, C, Pancotto, L, Ferlenghi, I, Scarselli, M, Pansegrau, W, Veggi, D, Merola, M, Cantini, F, Ruggiero, P, Banci, L, Masignani, V. | | Deposit date: | 2010-10-11 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Streptococcus pneumoniae RrgB pilus backbone D1 domain: in vivo protection and implications for the molecular mechanisms of pilus polymerization
To be Published
|
|
1TL5
 
 | | Solution structure of apoHAH1 | | Descriptor: | Copper transport protein ATOX1 | | Authors: | Anastassopoulou, I, Banci, L, Bertini, I, Cantini, F, Katsari, E, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-06-09 | | Release date: | 2004-10-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Apo and Copper(I)-Loaded Human Metallochaperone HAH1.
Biochemistry, 43, 2004
|
|
1TL4
 
 | | Solution structure of Cu(I) HAH1 | | Descriptor: | COPPER (I) ION, Copper transport protein ATOX1 | | Authors: | Anastassopoulou, I, Banci, L, Bertini, I, Cantini, F, Katsari, E, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-06-09 | | Release date: | 2004-10-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Apo and Copper(I)-Loaded Human Metallochaperone HAH1.
Biochemistry, 43, 2004
|
|
2MPF
 
 | | Solution structure human HCN2 CNBD in the cAMP-unbound state | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Saponaro, A, Pauleta, S.R, Cantini, F, Matzapetakis, M, Hammann, C, Banci, L, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2014-05-16 | | Release date: | 2014-09-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the mutual antagonism of cAMP and TRIP8b in regulating HCN channel function.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2NCL
 
 | |
1F22
 
 | | A PROTON-NMR INVESTIGATION OF THE FULLY REDUCED CYTOCHROME C7 FROM DESULFUROMONAS ACETOXIDANS. COMPARISON BETWEEN THE REDUCED AND THE OXIDIZED FORMS. | | Descriptor: | CYTOCHROME C7, HEME C | | Authors: | Assfalg, M, Banci, L, Bertini, I, Bruschi, M, Giudici-Orticoni, M.T. | | Deposit date: | 2000-05-23 | | Release date: | 2000-06-21 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A proton-NMR investigation of the fully reduced cytochrome c7 from Desulfuromonas acetoxidans. Comparison between the reduced and the oxidized forms.
Eur.J.Biochem., 266, 1999
|
|
1J6Q
 
 | | Solution structure and characterization of the heme chaperone CcmE | | Descriptor: | cytochrome c maturation protein E | | Authors: | Arnesano, F, Banci, L, Barker, P.D, Bertini, I, Rosato, A, Su, X.C, Viezzoli, M.S. | | Deposit date: | 2002-04-30 | | Release date: | 2002-12-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and characterization of the heme chaperone CcmE
Biochemistry, 41, 2002
|
|
