6HOJ
 
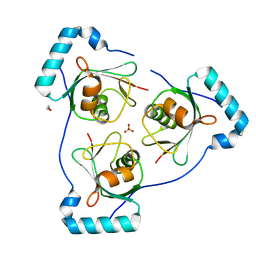 | | Structure of Beclin1 LIR motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, Beclin-1,Gamma-aminobutyric acid receptor-associated protein, SULFATE ION | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
2MUI
 
 | |
1UV6
 
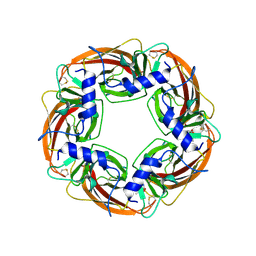 | | X-ray structure of acetylcholine binding protein (AChBP) in complex with carbamylcholine | | Descriptor: | 2-[(AMINOCARBONYL)OXY]-N,N,N-TRIMETHYLETHANAMINIUM, ACETYLCHOLINE-BINDING PROTEIN | | Authors: | Celie, P.H.N, Van Rossum-fikkert, S.E, Van Dijk, W.J, Brejc, K, Smit, A.B, Sixma, T.K. | | Deposit date: | 2004-01-15 | | Release date: | 2004-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nicotine and Carbamylcholine Binding to Nicotinic Acetylcholine Receptors as Studied in Achbp Crystal Structures
Neuron, 41, 2004
|
|
4U4S
 
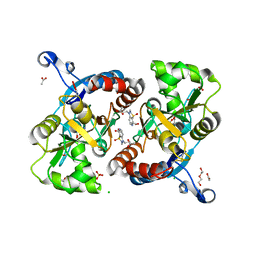 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and BPAM25 at 1.90 A resolution. | | Descriptor: | 4-ethyl-3,4-dihydro-2H-pyrido[4,3-e][1,2,4]thiadiazine 1,1-dioxide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Noerholm, A.B, Deva, T, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2014-07-24 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Positive Allosteric Modulators of 2-Amino-3-(3-hydroxy-5-methylisoxazol-4-yl)propionic Acid Receptors Belonging to 4-Cyclopropyl-3,4-dihydro-2H-1,2,4-pyridothiadiazine Dioxides and Diversely Chloro-Substituted 4-Cyclopropyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-Dioxides.
J.Med.Chem., 57, 2014
|
|
4U4X
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and BPAM37 at 1.56 A resolution. | | Descriptor: | 4-ethyl-3,4-dihydro-2H-pyrido[3,2-e][1,2,4]thiadiazine 1,1-dioxide, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Noerholm, A.B, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2014-07-24 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Positive Allosteric Modulators of 2-Amino-3-(3-hydroxy-5-methylisoxazol-4-yl)propionic Acid Receptors Belonging to 4-Cyclopropyl-3,4-dihydro-2H-1,2,4-pyridothiadiazine Dioxides and Diversely Chloro-Substituted 4-Cyclopropyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-Dioxides.
J.Med.Chem., 57, 2014
|
|
1UW6
 
 | | X-ray structure of acetylcholine binding protein (AChBP) in complex with nicotine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, ACETYLCHOLINE-BINDING PROTEIN | | Authors: | Celie, P.H.N, Van Rossum-fikkert, S.E, Van Dijk, W.J, Brejc, K, Smit, A.B, Sixma, T.K. | | Deposit date: | 2004-01-30 | | Release date: | 2004-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nicotine and Carbamylcholine Binding to Nicotinic Acetylcholine Receptors as Studied in Achbp Crystal Structures
Neuron, 41, 2004
|
|
1UX2
 
 | | X-ray structure of acetylcholine binding protein (AChBP) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYLCHOLINE BINDING PROTEIN, ... | | Authors: | Celie, P.H.N, Van Rossum-fikkert, S.E, Van Dijk, W.J, Brejc, K, Smit, A.B, Sixma, T.K. | | Deposit date: | 2004-02-18 | | Release date: | 2004-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nicotine and Carbamylcholine Binding to Nicotinic Acetylcholine Receptors as Studied in Achbp Crystal Structures
Neuron, 41, 2004
|
|
6X9V
 
 | | HIV-1 Envelope Glycoprotein BG505 SOSIP.664, expressed in HEK293S cells and deglycosylated by endoglycosidase H, in complex with RM20A3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Envelope Glycoprotein BG505 SOSIP.664 gp120, ... | | Authors: | Berndsen, Z.T, Ward, A.B. | | Deposit date: | 2020-06-03 | | Release date: | 2020-11-04 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Visualization of the HIV-1 Env glycan shield across scales.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XBS
 
 | | Streptomyces coelicolor methylmalonyl-CoA epimerase (E43Q) in complex with 2-nitronate-propionyl-CoA | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Methylmalonyl-CoA epimerase, ... | | Authors: | Stunkard, L.M, Boram, T.J, Benjamin, A.B, Bower, J.B, Lohman, J.R. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Streptomyces coelicolor methylmalonyl-CoA epimerase (E43Q) in complex with 2-nitronate-propionyl-CoA
To Be Published
|
|
6XSZ
 
 | |
6X67
 
 | | Cryo-EM structure of piggyBac transposase strand transfer complex (STC) | | Descriptor: | CALCIUM ION, DNA (37-MER), DNA (47-MER), ... | | Authors: | Chen, Q, Hickman, A.B, Dyda, F. | | Deposit date: | 2020-05-27 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis of seamless excision and specific targeting by piggyBac transposase
Nat Commun, 11, 2020
|
|
6XBQ
 
 | | Streptomyces coelicolor methylmalonyl-CoA epimerase in complex with carboxy-carba(dethia)-CoA | | Descriptor: | CARBOXYMETHYLDETHIA COENZYME *A, COBALT (II) ION, Methylmalonyl-CoA epimerase, ... | | Authors: | Stunkard, L.M, Boram, T.J, Benjamin, A.B, Bower, J.B, Lohman, J.R. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Streptomyces coelicolor methylmalonyl-CoA epimerase in complex with carboxy-carba(dethia)-CoA
To Be Published
|
|
6XBT
 
 | | Streptomyces coelicolor methylmalonyl-CoA epimerase (Q60A) in complex with 2-nitronate-propionyl-CoA | | Descriptor: | COBALT (II) ION, COENZYME A, Methylmalonyl-CoA epimerase, ... | | Authors: | Stunkard, L.M, Boram, T.J, Benjamin, A.B, Bower, J.B, Lohman, J.R. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Streptomyces coelicolor methylmalonyl-CoA epimerase (Q60A) in complex with 2-nitronate-propionyl-CoA
To Be Published
|
|
6X9T
 
 | |
6XBR
 
 | | Streptomyces coelicolor methylmalonyl-CoA epimerase (E43L) in complex with 2-nitronate-propionyl-CoA | | Descriptor: | AMMONIUM ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Stunkard, L.M, Boram, T.J, Benjamin, A.B, Bower, J.B, Lohman, J.R. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Streptomyces coelicolor methylmalonyl-CoA epimerase (E43L) in complex with 2-nitronate-propionyl-CoA
To Be Published
|
|
6XBV
 
 | | Streptomyces coelicolor methylmalonyl-CoA epimerase (S115T) in complex with 2-nitronate-propionyl-CoA | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Methylmalonyl-CoA epimerase, ... | | Authors: | Stunkard, L.M, Boram, T.J, Benjamin, A.B, Bower, J.B, Lohman, J.R. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Streptomyces coelicolor methylmalonyl-CoA epimerase (S115T) in complex with 2-nitronate-propionyl-CoA
To Be Published
|
|
6XX1
 
 | | The unique CBM3-Clocl_1192 of Hungateiclostridium clariflavum | | Descriptor: | Cellulose binding domain-containing protein | | Authors: | Milana, M.V, Almog, R, Yaniv, O, Oded, L, Inna, R.G, Felix, F, Edward, A.B, Raphael, L. | | Deposit date: | 2020-01-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The unique CBM3-Cthe_0271 from Ruminoclostridium thermocellum and CBM3-Clocl_1192 from Hungateiclostridium clariflavum
To Be Published
|
|
6XT1
 
 | |
7N5F
 
 | | Structure of Mechanosensitive Ion Channel Flycatcher1 Protomer in 'Down' conformation in GDN | | Descriptor: | Mechanosensitive ion channel Flycatcher1, PALMITIC ACID | | Authors: | Jojoa-Cruz, S, Saotome, K, Lee, W.H, Patapoutian, A, Ward, A.B. | | Deposit date: | 2021-06-05 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into the Venus flytrap mechanosensitive ion channel Flycatcher1.
Nat Commun, 13, 2022
|
|
7N5D
 
 | | Composite Structure of Mechanosensitive Ion Channel Flycatcher1 in GDN | | Descriptor: | Mechanosensitive ion channel Flycatcher1, PALMITIC ACID | | Authors: | Jojoa-Cruz, S, Saotome, K, Lee, W.H, Patapoutian, A, Ward, A.B. | | Deposit date: | 2021-06-05 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into the Venus flytrap mechanosensitive ion channel Flycatcher1.
Nat Commun, 13, 2022
|
|
7N5G
 
 | | Structure of Mechanosensitive Ion Channel Flycatcher1 Protomer in 'Up' conformation in GDN | | Descriptor: | Mechanosensitive ion channel Flycatcher1, PALMITIC ACID | | Authors: | Jojoa-Cruz, S, Saotome, K, Lee, W.H, Patapoutian, A, Ward, A.B. | | Deposit date: | 2021-06-05 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural insights into the Venus flytrap mechanosensitive ion channel Flycatcher1.
Nat Commun, 13, 2022
|
|
7N5E
 
 | | Structure of Mechanosensitive Ion Channel Flycatcher1 in GDN | | Descriptor: | Mechanosensitive ion channel Flycatcher1, PALMITIC ACID | | Authors: | Jojoa-Cruz, S, Saotome, K, Lee, W.H, Patapoutian, A, Ward, A.B. | | Deposit date: | 2021-06-05 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into the Venus flytrap mechanosensitive ion channel Flycatcher1.
Nat Commun, 13, 2022
|
|
6XSX
 
 | |
7O5H
 
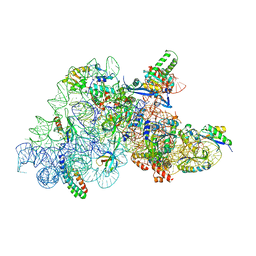 | | Ribosomal methyltransferase KsgA bound to small ribosomal subunit | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Stephan, N.C, Ries, A.B, Boehringer, D, Ban, N. | | Deposit date: | 2021-04-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of successive adenosine modifications by the conserved ribosomal methyltransferase KsgA.
Nucleic Acids Res., 49, 2021
|
|
5W9N
 
 | |
