3CUC
 
 | |
3CJL
 
 | |
3CK1
 
 | |
3D82
 
 | |
3DTT
 
 | |
3E0Z
 
 | |
3DUK
 
 | |
3DZA
 
 | |
3E10
 
 | |
3F9S
 
 | |
3FLJ
 
 | |
3F8X
 
 | |
3FGV
 
 | |
3CMB
 
 | | Crystal structure of acetoacetate decarboxylase (YP_001047042.1) from Methanoculleus marisnigri JR1 at 1.60 A resolution | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Acetoacetate decarboxylase, ... | | Authors: | Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-03-21 | | Release date: | 2008-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of acetoacetate decarboxylase (YP_001047042.1) from Methanoculleus marisnigri JR1 at 1.60 A resolution
To be published
|
|
3CT8
 
 | |
3CU3
 
 | |
3D5P
 
 | |
3CVJ
 
 | |
3D4E
 
 | |
3CJE
 
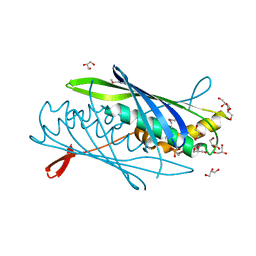 | |
3CEA
 
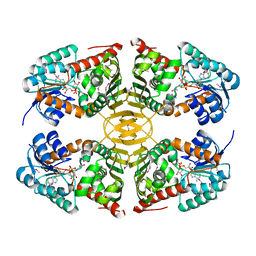 | |
3CIN
 
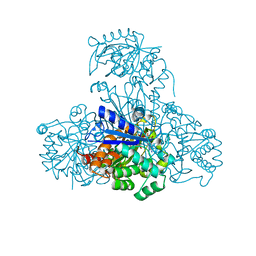 | |
3CJD
 
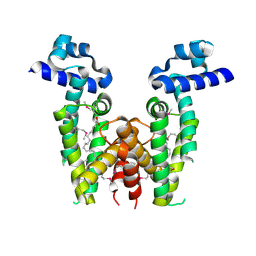 | |
3CNH
 
 | |
3CNX
 
 | |
