5ZPJ
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by phenylethylamine at pH 6 at 288 K (1) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Phenylethylamine oxidase, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZP6
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 6 at 277 K (2) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Phenylethylamine oxidase, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.688 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZPK
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by phenylethylamine at pH 6 at 288 K (2) | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZPG
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH6 at 293K (1) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Phenylethylamine oxidase, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZOY
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 7 at 288 K (2) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Phenylethylamine oxidase, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZPA
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 6 at 283 K (2) | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.689 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZPP
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by phenylethylamine at pH 8 at 288 K (3) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, PHENYLACETALDEHYDE, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
2YX9
 
 | | Crystal structure of D298K copper amine oxidase from Arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Murakawa, T, Okajima, T, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2007-04-25 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Trapping of a dopaquinone intermediate in the TPQ cofactor biogenesis in a copper-containing amine oxidase from Arthrobacter globiformis.
J.Am.Chem.Soc., 129, 2007
|
|
3VR2
 
 | | Crystal structure of nucleotide-free A3B3 complex from Enterococcus hirae V-ATPase [eA3B3] | | Descriptor: | V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | Authors: | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3VR4
 
 | | Crystal structure of Enterococcus hirae V1-ATPase [eV1] | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Saijo, S, Arai, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.172 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3VR6
 
 | | Crystal structure of AMP-PNP bound Enterococcus hirae V1-ATPase [bV1] | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, V-type sodium ATPase catalytic subunit A, ... | | Authors: | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3VR3
 
 | | Crystal structure of AMP-PNP bound A3B3 complex from Enterococcus hirae V-ATPase [bA3B3] | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, V-type sodium ATPase catalytic subunit A, ... | | Authors: | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
2ZFH
 
 | |
3VR5
 
 | | Crystal structure of nucleotide-free Enterococcus hirae V1-ATPase [eV1(L)] | | Descriptor: | V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B, V-type sodium ATPase subunit D, ... | | Authors: | Saijo, S, Arai, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3WA3
 
 | | Crystal structure of copper amine oxidase from arthrobacter globiformis in N2 condition | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Murakawa, T, Hayashi, H, Sunami, T, Kurihara, K, Tamada, T, Kuroki, R, Suzuki, M, Tanizawa, K, Okajima, T. | | Deposit date: | 2013-04-22 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High-resolution crystal structure of copper amine oxidase from Arthrobacter globiformis: assignment of bound diatomic molecules as O2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3WA2
 
 | | High resolution crystal structure of copper amine oxidase from arthrobacter globiformis | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Murakawa, T, Hayashi, H, Sunami, T, Kurihara, K, Tamada, T, Kuroki, R, Suzuki, M, Tanizawa, K, Okajima, T. | | Deposit date: | 2013-04-22 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | High-resolution crystal structure of copper amine oxidase from Arthrobacter globiformis: assignment of bound diatomic molecules as O2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3AMO
 
 | | Time-resolved X-ray Crystal Structure Analysis of Enzymatic Reaction of Copper Amine Oxidase from Arthrobacter globiformis | | Descriptor: | COPPER (II) ION, GLYCEROL, Phenylethylamine oxidase, ... | | Authors: | Kataoka, M, Oya, H, Tominaga, A, Otsu, M, Okajima, T, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2010-08-20 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Detection of the reaction intermediates catalyzed by a copper amine oxidase.
J.SYNCHROTRON RADIAT., 18, 2011
|
|
2ZPY
 
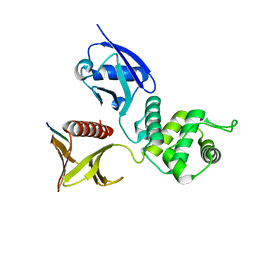 | | Crystal structure of the mouse radxin FERM domain complexed with the mouse CD44 cytoplasmic peptide | | Descriptor: | CD44 antigen, Radixin | | Authors: | Mori, T, Kitano, K, Terawaki, S, Maesaki, R, Fukami, Y, Hakoshima, T. | | Deposit date: | 2008-07-31 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for CD44 recognition by ERM proteins
J.Biol.Chem., 283, 2008
|
|
4BGN
 
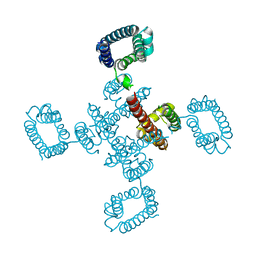 | | cryo-EM structure of the NavCt voltage-gated sodium channel | | Descriptor: | VOLTAGE-GATED SODIUM CHANNEL | | Authors: | Tsai, C.J, Tani, K, Irie, K, Hiroaki, Y, Shimomura, T, Mcmillan, D.G, Cook, G.M, Schertler, G, Fujiyoshi, Y, Li, X.D. | | Deposit date: | 2013-03-28 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | ELECTRON CRYSTALLOGRAPHY (9 Å) | | Cite: | Two Alternative Conformations of a Voltage-Gated Sodium Channel.
J.Mol.Biol., 425, 2013
|
|
3WE2
 
 | |
3WE3
 
 | |
4AOE
 
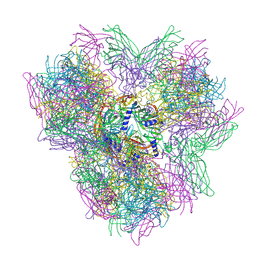 | | Biomphalaria glabrata Acetylcholine-binding protein type 2 (BgAChBP2) | | Descriptor: | ACETYLCHOLINE-BINDING PROTEIN TYPE 2 | | Authors: | Saur, M, Moeller, V, Kapetanopoulos, K, Braukmann, S, Gebauer, W, Tenzer, S, Markl, J. | | Deposit date: | 2012-03-26 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Acetylcholine-Binding Protein in the Hemolymph of the Planorbid Snail Biomphalaria Glabrata is a Pentagonal Dodecahedron (60 Subunits)
Plos One, 7, 2012
|
|
3X40
 
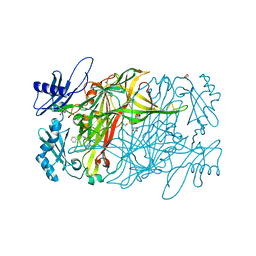 | | Copper amine oxidase from Arthrobacter globiformis: Product Schiff-base form produced by anaerobic reduction in the presence of sodium chloride | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2015-03-10 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
4AOD
 
 | | Biomphalaria glabrata Acetylcholine-binding protein type 1 (BgAChBP1) | | Descriptor: | ACETYLCHOLINE-BINDING PROTEIN TYPE 1 | | Authors: | Saur, M, Moeller, V, Kapetanopoulos, K, Braukmann, S, Gebauer, W, Tenzer, S, Markl, J. | | Deposit date: | 2012-03-26 | | Release date: | 2012-08-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Acetylcholine-Binding Protein in the Hemolymph of the Planorbid Snail Biomphalaria Glabrata is a Pentagonal Dodecahedron (60 Subunits)
Plos One, 7, 2012
|
|
3X3Y
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by histamine | | Descriptor: | COPPER (II) ION, GLYCEROL, POTASSIUM ION, ... | | Authors: | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2015-03-10 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
